Long non-coding RNA SNHG9 regulates viral replication in rhabdomyosarcoma cells infected with enterovirus D68 via miR-150-5p/c-Fos axis
- PMID: 36741904
- PMCID: PMC9893417
- DOI: 10.3389/fmicb.2022.1081237
Long non-coding RNA SNHG9 regulates viral replication in rhabdomyosarcoma cells infected with enterovirus D68 via miR-150-5p/c-Fos axis
Abstract
Background: The Enterovirus D68 (EV-D68) epidemic has increased knowledge of the virus as a pathogen capable of causing serious respiratory and neurological illnesses. It has been shown that long noncoding RNAs (lncRNAs) regulate viral replication and infection via multiple mechanisms or signaling pathways. However, the precise function of lncRNAs in EV-D68 infection remains unknown.
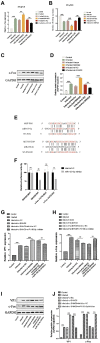
Methods: The differential expression profiles of lncRNA in EV-D68-infected and uninfected rhabdomyosarcoma (RD) cells were studied using high-throughput sequencing technology. The knockdown through small interfering RNA (siRNA) and overexpression of lncRNA SNHG9 (small ribonucleic acid host gene 9) were applied to investigate how lncRNA SNHG9 regulates EV-D68 propagation. The targeted interactions of lncRNA SNHG9 with miR-150-5p and miR-150-5p with c-Fos were validated using dual luciferase reporter system. LncRNA SNHG9 knockdown and miR-150-5p inhibitor were co-transfected with RD cells. QRT-PCR and western blot were used to detect RNA and protein levels, of c-Fos and VP1, respectively. Median tissue culture infectious dose (TCID50) was applied to detect viral titers.
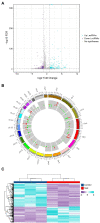
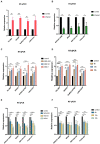
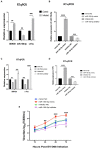
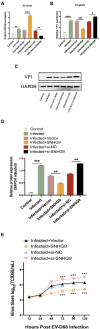
Results: The results demonstrated that a total of 375 lncRNAs were highly dysregulated in the EV-D68 infection model. In the EV-D68 infection model, lncRNA SNHG9 and c-Fos were increased in EV-D68-infected RD cells. However, the expression level of miR-150-5p was downregulated. In addition, overexpression of SNHG9 in RD cells resulted in decreased viral replication levels and viral titers following infection with EV-D68, and further experiments revealed that overexpression of SNHG9 inhibited the viral replication by targeting increased miR-150-5p binding and significantly increased c-Fos expression in RD cells.
Conclusion: Our findings indicate that the SNHG9/miR-150-5p/c-Fos axis influences EV-D68 replication in host cells and that SNHG9 may be a possible target for anti-EV-D68 infection therapies.
Keywords: SNHG9; c-Fos; ceRNA; enterovirus D68; infection; miR-150-5p.
Copyright © 2023 Fu, Si, Xu, Tang, He, Lu, Li, Li, Gao and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
High throughput sequencing of whole transcriptome and construct of ceRNA regulatory network in RD cells infected with enterovirus D68.Virol J. 2021 Nov 7;18(1):216. doi: 10.1186/s12985-021-01686-x. Virol J. 2021. PMID: 34743709 Free PMC article.
-
ADAR1p110 promotes Enterovirus D68 replication through its deaminase domain and inhibition of PKR pathway.Virol J. 2022 Dec 22;19(1):222. doi: 10.1186/s12985-022-01952-6. Virol J. 2022. PMID: 36550502 Free PMC article.
-
Susceptibility of Enterovirus-D68 to RNAi-mediated antiviral knockdown.Antiviral Res. 2019 Oct;170:104565. doi: 10.1016/j.antiviral.2019.104565. Epub 2019 Jul 20. Antiviral Res. 2019. PMID: 31336148
-
SNHG9/miR-199a-5p/Wnt2 Axis Regulates Cell Growth and Aerobic Glycolysis in Glioblastoma.J Neuropathol Exp Neurol. 2019 Oct 1;78(10):939-948. doi: 10.1093/jnen/nlz078. J Neuropathol Exp Neurol. 2019. PMID: 31504670
-
Enterovirus D68 Antivirals: Past, Present, and Future.ACS Infect Dis. 2020 Jul 10;6(7):1572-1586. doi: 10.1021/acsinfecdis.0c00120. Epub 2020 May 14. ACS Infect Dis. 2020. PMID: 32352280 Free PMC article. Review.
Cited by
-
Multidisciplinary advances in kombucha fermentation, health efficacy, and market evolution.Arch Microbiol. 2024 Aug 5;206(9):366. doi: 10.1007/s00203-024-04086-1. Arch Microbiol. 2024. PMID: 39098983 Review.
References
-
- Aznaourova M., Schmerer N., Janga H., Zhang Z., Pauck K., Bushe J., et al. . (2022). Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proc. Natl. Acad. Sci. U. S. A. 119:e2120680119. doi: 10.1073/pnas.2120680119, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

