The emerging landscape of novel 4-1BB (CD137) agonistic drugs for cancer immunotherapy
- PMID: 36727218
- PMCID: PMC9897756
- DOI: 10.1080/19420862.2023.2167189
The emerging landscape of novel 4-1BB (CD137) agonistic drugs for cancer immunotherapy
Abstract
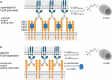
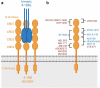
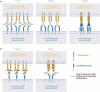
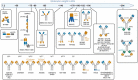
The clinical development of 4-1BB agonists for cancer immunotherapy has raised substantial interest during the past decade. The first generation of 4-1BB agonistic antibodies entering the clinic, urelumab (BMS-663513) and utomilumab (PF-05082566), failed due to (liver) toxicity or lack of efficacy, respectively. The two antibodies display differences in the affinity and the 4-1BB receptor epitope recognition, as well as the isotype, which determines the Fc-gamma-receptor (FcγR) crosslinking activity. Based on this experience a very diverse landscape of second-generation 4-1BB agonists addressing the liabilities of first-generation agonists has recently been developed, with many entering clinical Phase 1 and 2 studies. This review provides an overview focusing on differences and their scientific rationale, as well as challenges foreseen during the clinical development of these molecules.
Keywords: 4-1BB; 4-1BB agonists; CD137; TNFRSF9; bispecific antibodies; cancer immunotherapy; costimulatory agonist.
Conflict of interest statement
All authors are Roche employees and declare ownership of Roche stock options. Authors are inventors on patent applications (WO2016075278, WO2018114754, and WO2018114748) held/submitted by F. Hoffmann La Roche AG that cover tumor-targeted 4-1BBLs and their combination therapy.
Figures



Similar articles
-
A humanized 4-1BB-targeting agonistic antibody exerts potent antitumor activity in colorectal cancer without systemic toxicity.J Transl Med. 2022 Sep 8;20(1):415. doi: 10.1186/s12967-022-03619-w. J Transl Med. 2022. PMID: 36076251 Free PMC article.
-
Optimization of 4-1BB antibody for cancer immunotherapy by balancing agonistic strength with FcγR affinity.Nat Commun. 2019 May 20;10(1):2141. doi: 10.1038/s41467-019-10088-1. Nat Commun. 2019. PMID: 31105267 Free PMC article.
-
Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies.Blood. 2018 Jan 4;131(1):49-57. doi: 10.1182/blood-2017-06-741041. Epub 2017 Nov 8. Blood. 2018. PMID: 29118009 Review.
-
Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.Nat Commun. 2018 Nov 8;9(1):4679. doi: 10.1038/s41467-018-07136-7. Nat Commun. 2018. PMID: 30410017 Free PMC article.
-
Targeting CD137 (4-1BB) towards improved safety and efficacy for cancer immunotherapy.Front Immunol. 2023 Jun 2;14:1208788. doi: 10.3389/fimmu.2023.1208788. eCollection 2023. Front Immunol. 2023. PMID: 37334375 Free PMC article. Review.
Cited by
-
Agonist Antibodies for Cancer Immunotherapy: History, Hopes, and Challenges.Clin Cancer Res. 2024 May 1;30(9):1712-1723. doi: 10.1158/1078-0432.CCR-23-1014. Clin Cancer Res. 2024. PMID: 38153346 Free PMC article. Review.
-
ATG-101 Is a Tetravalent PD-L1×4-1BB Bispecific Antibody That Stimulates Antitumor Immunity through PD-L1 Blockade and PD-L1-Directed 4-1BB Activation.Cancer Res. 2024 May 15;84(10):1680-1698. doi: 10.1158/0008-5472.CAN-23-2701. Cancer Res. 2024. PMID: 38501978 Free PMC article.
-
4-1BB immunotherapy: advances and hurdles.Exp Mol Med. 2024 Feb;56(1):32-39. doi: 10.1038/s12276-023-01136-4. Epub 2024 Jan 4. Exp Mol Med. 2024. PMID: 38172595 Free PMC article. Review.
-
Cancer therapy with antibodies.Nat Rev Cancer. 2024 Jun;24(6):399-426. doi: 10.1038/s41568-024-00690-x. Epub 2024 May 13. Nat Rev Cancer. 2024. PMID: 38740967 Free PMC article. Review.
-
The present and future of bispecific antibodies for cancer therapy.Nat Rev Drug Discov. 2024 Apr;23(4):301-319. doi: 10.1038/s41573-024-00896-6. Epub 2024 Mar 6. Nat Rev Drug Discov. 2024. PMID: 38448606 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
