Protein post-translational modification in SARS-CoV-2 and host interaction
- PMID: 36713387
- PMCID: PMC9880545
- DOI: 10.3389/fimmu.2022.1068449
Protein post-translational modification in SARS-CoV-2 and host interaction
Abstract
SARS-CoV-2 can cause lung diseases, such as pneumonia and acute respiratory distress syndrome, and multi-system dysfunction. Post-translational modifications (PTMs) related to SARS-CoV-2 are conservative and pathogenic, and the common PTMs are glycosylation, phosphorylation, and acylation. The glycosylation of SARS-CoV-2 mainly occurs on spike (S) protein, which mediates the entry of the virus into cells through interaction with angiotensin-converting enzyme 2. SARS-CoV-2 utilizes glycans to cover its epitopes and evade the immune response through glycosylation of S protein. Phosphorylation of SARS-CoV-2 nucleocapsid (N) protein improves its selective binding to viral RNA and promotes viral replication and transcription, thereby increasing the load of the virus in the host. Succinylated N and membrane(M) proteins of SARS-CoV-2 synergistically affect virus particle assembly. N protein regulates its affinity for other proteins and the viral genome through acetylation. The acetylated envelope (E) protein of SARS-CoV-2 interacts with bromodomain-containing protein 2/4 to influence the host immune response. Both palmitoylation and myristoylation sites on S protein can affect the virus infectivity. Papain-like protease is a domain of NSP3 that dysregulates host inflammation by deubiquitination and impinges host IFN-I antiviral immune responses by deISGylation. Ubiquitination of ORF7a inhibits host IFN-α signaling by blocking STAT2 phosphorylation. The methylation of N protein can inhibit the formation of host stress granules and promote the binding of N protein to viral RNA, thereby promoting the production of virus particles. NSP3 macrodomain can reverse the ADP-ribosylation of host proteins, and inhibit the cascade immune response with IFN as the core, thereby promoting the intracellular replication of SARS-CoV-2. On the whole, PTMs have fundamental roles in virus entry, replication, particle assembly, and host immune response. Mutations in various SARS-CoV-2 variants, which lead to changes in PTMs at corresponding sites, cause different biological effects. In this paper, we mainly reviewed the effects of PTMs on SARS-CoV-2 and host cells, whose application is to inform the strategies for inhibiting viral infection and facilitating antiviral treatment and vaccine development for COVID-19.
Keywords: ADP-ribosylation; SARS-CoV-2; acylation; glycosylation; methylation; phosphorylation; ubiquitination.
Copyright © 2023 Cheng, Liu, Li, Sun, Liu, Wang, Shi and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
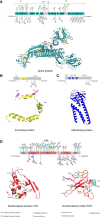
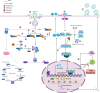
Similar articles
-
ISGylation of the SARS-CoV-2 N protein by HERC5 impedes N oligomerization and thereby viral RNA synthesis.J Virol. 2024 Sep 17;98(9):e0086924. doi: 10.1128/jvi.00869-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194248
-
Angiotensin-Converting Enzyme 2 Potentiates SARS-CoV-2 Infection by Antagonizing Type I Interferon Induction and Its Down-Stream Signaling Pathway.mSphere. 2022 Aug 31;7(4):e0021122. doi: 10.1128/msphere.00211-22. Epub 2022 Jul 12. mSphere. 2022. PMID: 35862802 Free PMC article.
-
Post-translational modifications of coronavirus proteins: roles and function.Future Virol. 2018 Jun;13(6):405-430. doi: 10.2217/fvl-2018-0008. Epub 2018 May 21. Future Virol. 2018. PMID: 32201497 Free PMC article. Review.
-
Therapeutic potential of green tea catechin, (-)-epigallocatechin-3-O-gallate (EGCG) in SARS-CoV-2 infection: Major interactions with host/virus proteases.Phytomed Plus. 2023 Feb;3(1):100402. doi: 10.1016/j.phyplu.2022.100402. Epub 2022 Dec 30. Phytomed Plus. 2023. PMID: 36597465 Free PMC article. Review.
-
SARS-CoV-2 Proteins: Are They Useful as Targets for COVID-19 Drugs and Vaccines?Curr Mol Med. 2022;22(1):50-66. doi: 10.2174/1566524021666210223143243. Curr Mol Med. 2022. PMID: 33622224 Review.
Cited by
-
Deciphering the Genetic Variation: A Comparative Analysis of Parental and Attenuated Strains of the QXL87 Vaccine for Infectious Bronchitis.Animals (Basel). 2024 Jun 13;14(12):1784. doi: 10.3390/ani14121784. Animals (Basel). 2024. PMID: 38929403 Free PMC article.
-
Phosphorylation of Human Polyomavirus Large and Small T Antigens: An Ignored Research Field.Viruses. 2023 Nov 9;15(11):2235. doi: 10.3390/v15112235. Viruses. 2023. PMID: 38005912 Free PMC article. Review.
-
Expanding horizons of iminosugars as broad-spectrum anti-virals: mechanism, efficacy and novel developments.Nat Prod Bioprospect. 2024 Sep 26;14(1):55. doi: 10.1007/s13659-024-00477-5. Nat Prod Bioprospect. 2024. PMID: 39325109 Free PMC article. Review.
-
Comprehensive genomic analysis of the SARS-CoV-2 Omicron variant BA.2.76 in Jining City, China, 2022.BMC Genomics. 2024 Apr 17;25(1):378. doi: 10.1186/s12864-024-10246-w. BMC Genomics. 2024. PMID: 38632523 Free PMC article.
-
Genetically predicted blood metabolites mediate the association between circulating immune cells and severe COVID-19: A Mendelian randomization study.Medicine (Baltimore). 2024 Nov 15;103(46):e40509. doi: 10.1097/MD.0000000000040509. Medicine (Baltimore). 2024. PMID: 39560514 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

