Centromeres as universal hotspots of DNA breakage, driving RAD51-mediated recombination during quiescence
- PMID: 36702125
- PMCID: PMC10009740
- DOI: 10.1016/j.molcel.2023.01.004
Centromeres as universal hotspots of DNA breakage, driving RAD51-mediated recombination during quiescence
Abstract
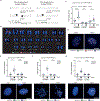
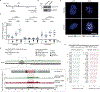
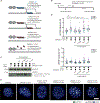
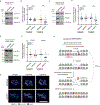
Centromeres are essential for chromosome segregation in most animals and plants yet are among the most rapidly evolving genome elements. The mechanisms underlying this paradoxical phenomenon remain enigmatic. Here, we report that human centromeres innately harbor a striking enrichment of DNA breaks within functionally active centromere regions. Establishing a single-cell imaging strategy that enables comparative assessment of DNA breaks at repetitive regions, we show that centromeric DNA breaks are induced not only during active cellular proliferation but also de novo during quiescence. Markedly, centromere DNA breaks in quiescent cells are resolved enzymatically by the evolutionarily conserved RAD51 recombinase, which in turn safeguards the specification of functional centromeres. This study highlights the innate fragility of centromeres, which may have been co-opted over time to reinforce centromere specification while driving rapid evolution. The findings also provide insights into how fragile centromeres are likely to contribute to human disease.
Keywords: CENP-A; DNA damage; RAD51; centromeres; homologous recombination.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans.PLoS Genet. 2014 Apr 24;10(4):e1004344. doi: 10.1371/journal.pgen.1004344. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24762765 Free PMC article.
-
Activation of homologous recombination in G1 preserves centromeric integrity.Nature. 2021 Dec;600(7890):748-753. doi: 10.1038/s41586-021-04200-z. Epub 2021 Dec 1. Nature. 2021. PMID: 34853474
-
Regulation of mitotic recombination between DNA repeats in centromeres.Nucleic Acids Res. 2017 Nov 2;45(19):11222-11235. doi: 10.1093/nar/gkx763. Nucleic Acids Res. 2017. PMID: 28977643 Free PMC article.
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
-
Centromeres Drive a Hard Bargain.Trends Genet. 2017 Feb;33(2):101-117. doi: 10.1016/j.tig.2016.12.001. Epub 2017 Jan 7. Trends Genet. 2017. PMID: 28069312 Free PMC article. Review.
Cited by
-
The complete diploid reference genome of RPE-1 identifies human phased epigenetic landscapes.bioRxiv [Preprint]. 2023 Dec 30:2023.11.01.565049. doi: 10.1101/2023.11.01.565049. bioRxiv. 2023. PMID: 38168337 Free PMC article. Preprint.
-
Genome maintenance meets mechanobiology.Chromosoma. 2024 Jan;133(1):15-36. doi: 10.1007/s00412-023-00807-5. Epub 2023 Aug 15. Chromosoma. 2024. PMID: 37581649 Free PMC article. Review.
-
Most large structural variants in cancer genomes can be detected without long reads.Nat Genet. 2023 Dec;55(12):2139-2148. doi: 10.1038/s41588-023-01540-6. Epub 2023 Nov 9. Nat Genet. 2023. PMID: 37945902 Free PMC article.
-
DNA Damage Atlas: an atlas of DNA damage and repair.Nucleic Acids Res. 2024 Jan 5;52(D1):D1218-D1226. doi: 10.1093/nar/gkad845. Nucleic Acids Res. 2024. PMID: 37831087 Free PMC article.
-
Expansion of human centromeric arrays in cells undergoing break-induced replication.bioRxiv [Preprint]. 2023 Nov 15:2023.11.11.566714. doi: 10.1101/2023.11.11.566714. bioRxiv. 2023. Update in: Cell Rep. 2024 Mar 26;43(3):113851. doi: 10.1016/j.celrep.2024.113851. PMID: 38014305 Free PMC article. Updated. Preprint.
References
-
- Willard HF, and Waye JS (1987). Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 25, 207–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

