The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation
- PMID: 36700827
- PMCID: PMC10952680
- DOI: 10.1002/1873-3468.14586
The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation
Abstract
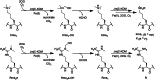
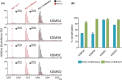
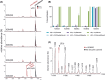
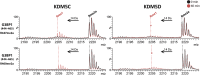
The demethylation of Nε -methyllysine residues on histones by Jumonji-C lysine demethylases (JmjC-KDMs) has been established. A subset of JmjC-KDMs has also been reported to have Nω -methylarginine residue demethylase (RDM) activity. Here, we describe biochemical screening studies, showing that the catalytic domains of all human KDM5s (KDM5A-KDM5D), KDM4E and, to a lesser extent, KDM4A/D, have both KDM and RDM activities with histone peptides. Ras GTPase-activating protein-binding protein 1 peptides were shown to be RDM substrates for KDM5C/D. No RDM activity was observed with KDM1A and the other JmjC-KDMs tested. The results highlight the potential of JmjC-KDMs to catalyse reactions other than Nε -methyllysine demethylation. Although our study is limited to peptide fragments, the results should help guide biological studies investigating JmjC functions.
Keywords: 2-oxoglutarate non-heme oxygenase; JmjC-KDM; epigenetics; histone N-methyl arginine/lysine demethylase; post translational modification.
© 2023 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

Similar articles
-
Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates.Epigenetics. 2014 Dec;9(12):1596-603. doi: 10.4161/15592294.2014.983381. Epigenetics. 2014. PMID: 25625844 Free PMC article.
-
Inhibitors of both the N-methyl lysyl- and arginyl-demethylase activities of the JmjC oxygenases.Philos Trans R Soc Lond B Biol Sci. 2018 Jun 5;373(1748):20170071. doi: 10.1098/rstb.2017.0071. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29685975 Free PMC article.
-
Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases.Nat Commun. 2016 Jun 23;7:11974. doi: 10.1038/ncomms11974. Nat Commun. 2016. PMID: 27337104 Free PMC article.
-
Recent developments in catalysis and inhibition of the Jumonji histone demethylases.Curr Opin Struct Biol. 2023 Dec;83:102707. doi: 10.1016/j.sbi.2023.102707. Epub 2023 Oct 11. Curr Opin Struct Biol. 2023. PMID: 37832177 Free PMC article. Review.
-
Taking Me away: the function of phosphorylation on histone lysine demethylases.Trends Biochem Sci. 2024 Mar;49(3):257-276. doi: 10.1016/j.tibs.2023.12.004. Epub 2024 Jan 16. Trends Biochem Sci. 2024. PMID: 38233282 Review.
Cited by
-
R-Methylation in Plants: A Key Regulator of Plant Development and Response to the Environment.Int J Mol Sci. 2024 Sep 14;25(18):9937. doi: 10.3390/ijms25189937. Int J Mol Sci. 2024. PMID: 39337424 Free PMC article. Review.
-
Domain architecture and protein-protein interactions regulate KDM5A recruitment to the chromatin.Epigenetics. 2023 Dec;18(1):2268813. doi: 10.1080/15592294.2023.2268813. Epub 2023 Oct 15. Epigenetics. 2023. PMID: 37838974 Free PMC article. Review.
-
High-throughput mRNA sequencing of human placenta shows sex differences across gestation.Placenta. 2024 May;150:8-21. doi: 10.1016/j.placenta.2024.03.005. Epub 2024 Mar 21. Placenta. 2024. PMID: 38537412
-
Versatile JMJD proteins: juggling histones and much more.Trends Biochem Sci. 2024 Sep;49(9):804-818. doi: 10.1016/j.tibs.2024.06.009. Epub 2024 Jun 26. Trends Biochem Sci. 2024. PMID: 38926050 Review.
References
-
- Allis CD and Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500. - PubMed
-
- Paik WK and Kim S (1973) Enzymatic demethylation of calf thymus histones. Biochem Biophys Res Commun 51, 781–788. - PubMed
-
- Markolovic S, Leissing TM, Chowdhury R, Wilkins SE, Lu X and Schofield CJ (2016) Structure‐function relationships of human JmjC oxygenases‐demethylases versus hydroxylases. Curr Opin Struct Biol 41, 62–72. - PubMed
-
- Forneris F, Binda C, Vanoni MA, Battaglioli E and Mattevi A (2005) Human histone demethylase LSD1 reads the histone code. J Biol Chem 280, 41360–41365. - PubMed
-
- Hopkinson RJ, Hamed RB, Rose NR, Claridge TD and Schofield CJ (2010) Monitoring the activity of 2‐oxoglutarate dependent histone demethylases by NMR spectroscopy: direct observation of formaldehyde. Chembiochem 11, 506–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

