RNA Pol III promoters-key players in precisely targeted plant genome editing
- PMID: 36685866
- PMCID: PMC9845283
- DOI: 10.3389/fgene.2022.989199
RNA Pol III promoters-key players in precisely targeted plant genome editing
Abstract
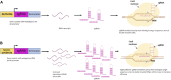
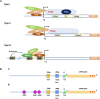
The clustered regularly interspaced short palindrome repeat (CRISPR)/CRISPR-associated protein Cas) system is a powerful and highly precise gene-editing tool in basic and applied research for crop improvement programs. CRISPR/Cas tool is being extensively used in plants to improve crop yield, quality, and nutritional value and make them tolerant to environmental stresses. CRISPR/Cas system consists of a Cas protein with DNA endonuclease activity and one CRISPR RNA transcript that is processed to form one or several short guide RNAs that direct Cas9 to the target DNA sequence. The expression levels of Cas proteins and gRNAs significantly influence the editing efficiency of CRISPR/Cas-mediated genome editing. This review focuses on insights into RNA Pol III promoters and their types that govern the expression levels of sgRNA in the CRISPR/Cas system. We discussed Pol III promoters structural and functional characteristics and their comparison with Pol II promoters. Further, the use of synthetic promoters to increase the targeting efficiency and overcome the structural, functional, and expressional limitations of RNA Pol III promoters has been discussed. Our review reports various studies that illustrate the use of endogenous U6/U3 promoters for improving editing efficiency in plants and the applicative approach of species-specific RNA pol III promoters for genome editing in model crops like Arabidopsis and tobacco, cereals, legumes, oilseed, and horticultural crops. We further highlight the significance of optimizing these species-specific promoters' systematic identification and validation for crop improvement and biotic and abiotic stress tolerance through CRISPR/Cas mediated genome editing.
Keywords: CRISPR/Cas9; RNA pol III promoters; TATA-box; U6 and U3 snRNA promoters; USE; synthetic promoter.
Copyright © 2023 Kor, Chowdhury, Keot, Yogendra, Chikkaputtaiah and Sudhakar Reddy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
In Planta Processing of the SpCas9-gRNA Complex.Plant Cell Physiol. 2017 Nov 1;58(11):1857-1867. doi: 10.1093/pcp/pcx154. Plant Cell Physiol. 2017. PMID: 29040704 Free PMC article.
-
Heterologous and endogenous U6 snRNA promoters enable CRISPR/Cas9 mediated genome editing in Aspergillus niger.Fungal Biol Biotechnol. 2018 Feb 8;5:2. doi: 10.1186/s40694-018-0047-4. eCollection 2018. Fungal Biol Biotechnol. 2018. PMID: 29456867 Free PMC article.
-
Functional characterization of Pol III U6 promoters for gene knockdown and knockout in Plutella xylostella.Insect Biochem Mol Biol. 2017 Oct;89:71-78. doi: 10.1016/j.ibmb.2017.08.009. Epub 2017 Sep 7. Insect Biochem Mol Biol. 2017. PMID: 28890398
-
CRISPR/Cas genome editing in soybean: challenges and new insights to overcome existing bottlenecks.J Adv Res. 2024 Aug 18:S2090-1232(24)00367-9. doi: 10.1016/j.jare.2024.08.024. Online ahead of print. J Adv Res. 2024. PMID: 39163906 Review.
-
CRISPR/Cas tool designs for multiplex genome editing and its applications in developing biotic and abiotic stress-resistant crop plants.Mol Biol Rep. 2022 Dec;49(12):11443-11467. doi: 10.1007/s11033-022-07741-2. Epub 2022 Aug 24. Mol Biol Rep. 2022. PMID: 36002653 Review.
Cited by
-
Optimization of CRISPR-Cas9 system in Eustoma grandiflorum.iScience. 2024 Feb 1;27(3):109053. doi: 10.1016/j.isci.2024.109053. eCollection 2024 Mar 15. iScience. 2024. PMID: 38361623 Free PMC article.
-
Strategies and Methods for Improving the Efficiency of CRISPR/Cas9 Gene Editing in Plant Molecular Breeding.Plants (Basel). 2023 Mar 28;12(7):1478. doi: 10.3390/plants12071478. Plants (Basel). 2023. PMID: 37050104 Free PMC article. Review.
-
Promoter Prediction in Agrobacterium tumefaciens Strain C58 by Using Artificial Intelligence Strategies.Methods Mol Biol. 2024;2844:33-44. doi: 10.1007/978-1-0716-4063-0_2. Methods Mol Biol. 2024. PMID: 39068330
-
Towards DNA-free CRISPR/Cas9 genome editing for sustainable oil palm improvement.3 Biotech. 2024 Jun;14(6):166. doi: 10.1007/s13205-024-04010-w. Epub 2024 May 28. 3 Biotech. 2024. PMID: 38817736 Review.
-
Plant Promoters: Their Identification, Characterization, and Role in Gene Regulation.Genes (Basel). 2023 Jun 6;14(6):1226. doi: 10.3390/genes14061226. Genes (Basel). 2023. PMID: 37372407 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

