Structural and evolutionary insights into astacin metallopeptidases
- PMID: 36685277
- PMCID: PMC9848320
- DOI: 10.3389/fmolb.2022.1080836
Structural and evolutionary insights into astacin metallopeptidases
Abstract
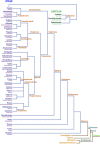
The astacins are a family of metallopeptidases (MPs) that has been extensively described from animals. They are multidomain extracellular proteins, which have a conserved core architecture encompassing a signal peptide for secretion, a prodomain or prosegment and a zinc-dependent catalytic domain (CD). This constellation is found in the archetypal name-giving digestive enzyme astacin from the European crayfish Astacus astacus. Astacin catalytic domains span ∼200 residues and consist of two subdomains that flank an extended active-site cleft. They share several structural elements including a long zinc-binding consensus sequence (HEXXHXXGXXH) immediately followed by an EXXRXDRD motif, which features a family-specific glutamate. In addition, a downstream SIMHY-motif encompasses a "Met-turn" methionine and a zinc-binding tyrosine. The overall architecture and some structural features of astacin catalytic domains match those of other more distantly related MPs, which together constitute the metzincin clan of metallopeptidases. We further analysed the structures of PRO-, MAM, TRAF, CUB and EGF-like domains, and described their essential molecular determinants. In addition, we investigated the distribution of astacins across kingdoms and their phylogenetic origin. Through extensive sequence searches we found astacin CDs in > 25,000 sequences down the tree of life from humans beyond Metazoa, including Choanoflagellata, Filasterea and Ichtyosporea. We also found < 400 sequences scattered across non-holozoan eukaryotes including some fungi and one virus, as well as in selected taxa of archaea and bacteria that are pathogens or colonizers of animal hosts, but not in plants. Overall, we propose that astacins originate in the root of Holozoa consistent with Darwinian descent and that the latter genes might be the result of horizontal gene transfer from holozoan donors.
Keywords: catalytic domain (CD); darwinian descent; evolution of metallopeptidases; horizontal gene transfer (HGT); phylogeny of enzymes.
Copyright © 2023 Gomis-Rüth and Stöcker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

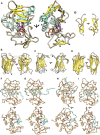


Similar articles
-
Functional and structural insights into astacin metallopeptidases.Biol Chem. 2012 Oct;393(10):1027-41. doi: 10.1515/hsz-2012-0149. Biol Chem. 2012. PMID: 23092796 Review.
-
Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.J Mol Biol. 1993 Feb 20;229(4):945-68. doi: 10.1006/jmbi.1993.1098. J Mol Biol. 1993. PMID: 8445658
-
Implications of the three-dimensional structure of astacin for the structure and function of the astacin family of zinc-endopeptidases.Eur J Biochem. 1993 May 15;214(1):215-31. doi: 10.1111/j.1432-1033.1993.tb17915.x. Eur J Biochem. 1993. PMID: 8508794
-
News from an ancient world: two novel astacin metalloproteases from the horseshoe crab.J Mol Biol. 2009 Jan 9;385(1):236-48. doi: 10.1016/j.jmb.2008.10.062. Epub 2008 Oct 30. J Mol Biol. 2009. PMID: 18996129
-
Structural aspects of the metzincin clan of metalloendopeptidases.Mol Biotechnol. 2003 Jun;24(2):157-202. doi: 10.1385/MB:24:2:157. Mol Biotechnol. 2003. PMID: 12746556 Review.
Cited by
-
Placozoan secretory cell types implicated in feeding, innate immunity and regulation of behavior.bioRxiv [Preprint]. 2025 Jan 16:2024.09.18.613768. doi: 10.1101/2024.09.18.613768. bioRxiv. 2025. PMID: 39372748 Free PMC article. Preprint.
-
Exploring oak processionary caterpillar induced lepidopterism (Part 1): unveiling molecular insights through transcriptomics and proteomics.Cell Mol Life Sci. 2024 Jul 27;81(1):311. doi: 10.1007/s00018-024-05330-z. Cell Mol Life Sci. 2024. PMID: 39066932 Free PMC article.
-
Salmon louse labial gland enzymes: implications for host settlement and immune modulation.Front Genet. 2024 Jan 17;14:1303898. doi: 10.3389/fgene.2023.1303898. eCollection 2023. Front Genet. 2024. PMID: 38299097 Free PMC article.
References
-
- AmbuAli A., Monaghan S. J., McLean K., Inglis N. F., Bekaert M., Wehner S., et al. (2020). Identification of proteins from the secretory/excretory products (SEPs) of the branchiuran ectoparasite Argulus foliaceus (Linnaeus, 1758) reveals unique secreted proteins amongst haematophagous ecdysozoa. Parasit. Vectors 13 (1), 88. 10.1186/s13071-020-3964-z - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

