Exosomal miRNA-profiling of pleural effusion in lung adenocarcinoma and tuberculosis
- PMID: 36684253
- PMCID: PMC9852630
- DOI: 10.3389/fsurg.2022.1050242
Exosomal miRNA-profiling of pleural effusion in lung adenocarcinoma and tuberculosis
Abstract
Background: Pleural effusion (PE) caused by lung cancer is prevalent, and it is difficult to differentiate it from PE caused by tuberculosis. Exosome-based liquid biopsy offers a non-invasive technique to diagnose benign and malignant PE. Exosomal miRNAs are potential diagnostic markers and play an essential role in signal transduction and biological processes in tumor development. We hypothesized that exosomal miRNA expression profiles in PE would contribute to identifying its diagnostic markers and elucidating the molecular basis of PE formation in lung cancer.
Methods: The exosomes from PE caused by lung adenocarcinoma (LUAD) and pulmonary tuberculosis were isolated and verified by transmission electron microscopy. The exosomal miRNA profiles were identified using deep sequencing and validated with quantitative real-time PCR (qRT-PCR). We performed bioinformatic analysis for differentially expressed miRNAs to explore how exosomal miRNAs regulate pleural effusion.
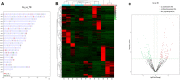
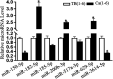
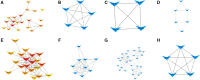
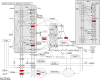
Results: We identified 99 upregulated and 91 downregulated miRNAs in malignant pleural effusion (MPE) compared to tuberculous pleural effusion (TPE). Seven differentially expressed miRNAs (DEmiRNAs) were validated by qRT-PCR, out of which 5 (71.4%) were confirmed through sequencing. Gene Ontology (GO) analysis revealed that most exosomal miRNAs target genes were involved in regulating cellular processes and nitrogen compound metabolism. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, the exosomal miRNAs target genes were mainly involved in Fc gamma R-mediated phagocytosis, Rap1 signaling pathway, and breast cancer. The hub genes, including ITGAM, FOXO1, MAPK14, YWHAB, GRIN1, and PRF1, were screened through plug-in cytoHubba. The PFR1 was identified as a critical gene in MPE formation using single-cell sequencing analysis. Additionally, we hypothesized that tumor cells affected natural killer cells and promoted the generation of PE in LUAD via the exosomal hsa-miR-3120-5p-PRF1 axis.
Conclusions: We identified exosomal miRNA profiles in LUAD-MPE and TPE, which may help in the differential diagnosis of MPE and TPE. Bioinformatic analysis revealed that these miRNAs might affect PE generation through tumor immune response in LUAD. Our results provided a new theoretical basis for understanding the function of exosomal miRNAs in LUAD-MPE.
Keywords: bioinformatic analysis; exosomes; lung adenocarcinoma; miRNAs; pleural effusions.
© 2023 Zhang, Bao, Yv and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
[Expression of circRNA_051778 in Lung Adenocarcinoma-Associated Malignant and Tuberculous Pleural Effusions and Its Clinical Significance].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 Sep 20;55(5):1254-1263. doi: 10.12182/20240960302. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39507963 Free PMC article. Chinese.
-
Integration of bioinformatics analysis and experimental validation identifies plasma exosomal miR-103b/877-5p/29c-5p as diagnostic biomarkers for early lung adenocarcinoma.Cancer Med. 2022 Dec;11(23):4411-4421. doi: 10.1002/cam4.4788. Epub 2022 May 18. Cancer Med. 2022. PMID: 35585716 Free PMC article.
-
Construction and analysis of expression profile of exosomal lncRNAs in pleural effusion in lung adenocarcinoma.J Clin Lab Anal. 2022 Dec;36(12):e24777. doi: 10.1002/jcla.24777. Epub 2022 Nov 25. J Clin Lab Anal. 2022. PMID: 36426920 Free PMC article.
-
Modulating Effects of Cancer-Derived Exosomal miRNAs and Exosomal Processing by Natural Products.Cancers (Basel). 2023 Jan 3;15(1):318. doi: 10.3390/cancers15010318. Cancers (Basel). 2023. PMID: 36612314 Free PMC article. Review.
-
Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis.Int J Mol Sci. 2023 Aug 4;24(15):12449. doi: 10.3390/ijms241512449. Int J Mol Sci. 2023. PMID: 37569824 Free PMC article. Review.
Cited by
-
Bladder Cancer in Exosomal Perspective: Unraveling New Regulatory Mechanisms.Int J Nanomedicine. 2024 Apr 22;19:3677-3695. doi: 10.2147/IJN.S458397. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38681092 Free PMC article. Review.
-
SOX9 promotes the invasion and migration of lung adenocarcinoma cells by activating the RAP1 signaling pathway.BMC Pulm Med. 2023 Nov 2;23(1):421. doi: 10.1186/s12890-023-02740-w. BMC Pulm Med. 2023. PMID: 37919693 Free PMC article.
-
Lung cancer cell-derived exosomes: progress on pivotal role and its application in diagnostic and therapeutic potential.Front Oncol. 2024 Oct 11;14:1459178. doi: 10.3389/fonc.2024.1459178. eCollection 2024. Front Oncol. 2024. PMID: 39464709 Free PMC article. Review.
-
The role of Mycobacterium tuberculosis exosomal miRNAs in host pathogen cross-talk as diagnostic and therapeutic biomarkers.Front Microbiol. 2024 Aug 8;15:1441781. doi: 10.3389/fmicb.2024.1441781. eCollection 2024. Front Microbiol. 2024. PMID: 39176271 Free PMC article. Review.
-
Non-coding RNAs in lung cancer: molecular mechanisms and clinical applications.Front Oncol. 2023 Sep 8;13:1256537. doi: 10.3389/fonc.2023.1256537. eCollection 2023. Front Oncol. 2023. PMID: 37746261 Free PMC article. Review.
References
-
- Buscail E, Alix-Panabières C, Quincy P, Cauvin T, Chauvet A, Degrandi O, et al. High clinical value of liquid biopsy to detect circulating tumor cells and tumor exosomes in pancreatic ductal adenocarcinoma patients eligible for up-front surgery. Cancers (Basel). (2019) 11(11):1656. 10.3390/cancers11111656 - DOI - PMC - PubMed
-
- Vitiello PP, De Falco V, Giunta EF, Ciardiello D, Cardone C, Vitale P, et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): a single institution experience. Cancers. (2019) 11(10):1504. 10.3390/cancers11101504 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

