Insight into the Global Negative Regulation of Iron Scavenger 7-HT Biosynthesis by the SigW/RsiW System in Pseudomonas donghuensis HYS
- PMID: 36674714
- PMCID: PMC9861184
- DOI: 10.3390/ijms24021184
Insight into the Global Negative Regulation of Iron Scavenger 7-HT Biosynthesis by the SigW/RsiW System in Pseudomonas donghuensis HYS
Abstract
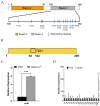
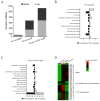
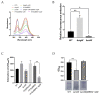
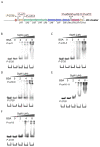
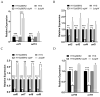
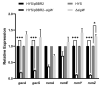
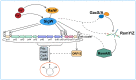
7-Hydroxytropolone (7-HT) is a unique iron scavenger synthesized by Pseudomonas donghuensis HYS that has various biological activities in addition to functioning as a siderophore. P. donghuensis HYS is more pathogenic than P. aeruginosa toward Caenorhabditis elegans, an observation that is closely linked to the biosynthesis of 7-HT. The nonfluorescent siderophore (nfs) gene cluster is responsible for the orderly biosynthesis of 7-HT and represents a competitive advantage that contributes to the increased survival of P. donghuensis HYS; however, the regulatory mechanisms of 7-HT biosynthesis remain unclear. This study is the first to propose that the ECF σ factor has a regulatory effect on 7-HT biosynthesis. In total, 20 ECF σ factors were identified through genome-wide scanning, and their responses to extracellular ferrous ions were characterized. We found that SigW was both significantly upregulated under high-iron conditions and repressed by an adjacent anti-σ factor. RNA-Seq results suggest that the SigW/RsiW system is involved in iron metabolism and 7-HT biosynthesis. Combined with the siderophore phenotype, we also found that SigW could inhibit siderophore synthesis, and this inhibition can be relieved by RsiW. EMSA assays proved that SigW, when highly expressed, can directly bind to the promoter region of five operons of the nfs cluster to inhibit the transcription of the corresponding genes and consequently suppress 7-HT biosynthesis. In addition, SigW not only directly negatively regulates structural genes related to 7-HT synthesis but also inhibits the transcription of regulatory proteins, including of the Gac/Rsm cascade system. Taken together, our results highlight that the biosynthesis of 7-HT is negatively regulated by SigW and that the SigW/RsiW system is involved in mechanisms for the regulation of iron homeostasis in P. donghuensis HYS. As a result of this work, we identified a novel mechanism for the global negative regulation of 7-HT biosynthesis, complementing our understanding of the function of ECF σ factors in Pseudomonas.
Keywords: 7-hydroxytropolone; Pseudomonas donghuensis HYS; SigW; anti-σ factor; negative regulator; siderophore.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
A Complex Mechanism Involving LysR and TetR/AcrR That Regulates Iron Scavenger Biosynthesis in Pseudomonas donghuensis HYS.J Bacteriol. 2018 Jun 11;200(13):e00087-18. doi: 10.1128/JB.00087-18. Print 2018 Jul 1. J Bacteriol. 2018. PMID: 29686142 Free PMC article.
-
The Gene paaZ of the Phenylacetic Acid (PAA) Catabolic Pathway Branching Point and ech outside the PAA Catabolon Gene Cluster Are Synergistically Involved in the Biosynthesis of the Iron Scavenger 7-Hydroxytropolone in Pseudomonas donghuensis HYS.Int J Mol Sci. 2023 Aug 10;24(16):12632. doi: 10.3390/ijms241612632. Int J Mol Sci. 2023. PMID: 37628812 Free PMC article.
-
Pseudomonas donghuensis HYS 7-hydroxytropolone contributes to pathogenicity toward Caenorhabditis elegans and is influenced by pantothenic acid.Biochem Biophys Res Commun. 2020 Nov 26;533(1):50-56. doi: 10.1016/j.bbrc.2020.08.067. Epub 2020 Sep 10. Biochem Biophys Res Commun. 2020. PMID: 32921415
-
Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas.Mol Microbiol. 2002 Sep;45(5):1177-90. doi: 10.1046/j.1365-2958.2002.03088.x. Mol Microbiol. 2002. PMID: 12207687 Review.
-
Gene regulation of siderophore-mediated iron acquisition in Pseudomonas: not only the Fur repressor.Mol Microbiol. 1995 Aug;17(4):603-10. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. Mol Microbiol. 1995. PMID: 8801415 Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

