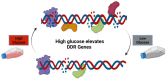
High Glucose Increases DNA Damage and Elevates the Expression of Multiple DDR Genes
- PMID: 36672885
- PMCID: PMC9858638
- DOI: 10.3390/genes14010144
High Glucose Increases DNA Damage and Elevates the Expression of Multiple DDR Genes
Abstract
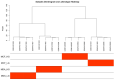
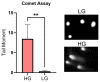
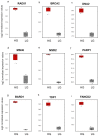
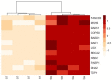
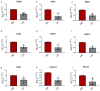
The DNA Damage Response (DDR) pathways sense DNA damage and coordinate robust DNA repair and bypass mechanisms. A series of repair proteins are recruited depending on the type of breaks and lesions to ensure overall survival. An increase in glucose levels was shown to induce genome instability, yet the links between DDR and glucose are still not well investigated. In this study, we aimed to identify dysregulation in the transcriptome of normal and cancerous breast cell lines upon changing glucose levels. We first performed bioinformatics analysis using a microarray dataset containing the triple-negative breast cancer (TNBC) MDA-MB-231 and the normal human mammary epithelium MCF10A cell lines grown in high glucose (HG) or in the presence of the glycolysis inhibitor 2-deoxyglucose (2DG). Interestingly, multiple DDR genes were significantly upregulated in both cell lines grown in HG. In the wet lab, we remarkably found that HG results in severe DNA damage to TNBC cells as observed using the comet assay. In addition, several DDR genes were confirmed to be upregulated using qPCR analysis in the same cell line. Our results propose a strong need for DDR pathways in the presence of HG to oppose the severe DNA damage induced in cells.
Keywords: DNA damage; DNA damage response (DDR); cancer; diabetes mellitus; hyperglycemia (HG); metabolic diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
[Application value of DNA damage repair variants in adjuvant therapy of triple negative breast cancer].Zhonghua Zhong Liu Za Zhi. 2023 Sep 23;45(9):787-795. doi: 10.3760/cma.j.cn112152-20220912-00612. Zhonghua Zhong Liu Za Zhi. 2023. PMID: 37805443 Chinese.
-
Chinese medicine formula 'Baipuhuang Keli' inhibits triple-negative breast cancer by hindering DNA damage repair via MAPK/ERK pathway.J Ethnopharmacol. 2023 Mar 25;304:116077. doi: 10.1016/j.jep.2022.116077. Epub 2022 Dec 24. J Ethnopharmacol. 2023. PMID: 36572327
-
Regorafenib sensitizes human breast cancer cells to radiation by inhibiting multiple kinases and inducing DNA damage.Int J Radiat Biol. 2021;97(8):1109-1120. doi: 10.1080/09553002.2020.1730012. Epub 2020 Mar 2. Int J Radiat Biol. 2021. PMID: 32052681 Free PMC article.
-
DNA damage response inhibitors: An avenue for TNBC treatment.Biochim Biophys Acta Rev Cancer. 2021 Apr;1875(2):188521. doi: 10.1016/j.bbcan.2021.188521. Epub 2021 Feb 5. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33556453 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
Special Issue "DNA Replication/Repair, and the DNA Damage Response in Human Disease".Genes (Basel). 2023 Apr 11;14(4):893. doi: 10.3390/genes14040893. Genes (Basel). 2023. PMID: 37107651 Free PMC article.
-
The "sweet" path to cancer: focus on cellular glucose metabolism.Front Oncol. 2023 May 25;13:1202093. doi: 10.3389/fonc.2023.1202093. eCollection 2023. Front Oncol. 2023. PMID: 37305566 Free PMC article. Review.
-
Repurposing metabolic regulators: antidiabetic drugs as anticancer agents.Mol Biomed. 2024 Sep 28;5(1):40. doi: 10.1186/s43556-024-00204-z. Mol Biomed. 2024. PMID: 39333445 Free PMC article. Review.
-
Diabetes and Cancer: A Twisted Bond.Oncol Rev. 2024 May 21;18:1354549. doi: 10.3389/or.2024.1354549. eCollection 2024. Oncol Rev. 2024. PMID: 38835644 Free PMC article. Review.
References
-
- Ingram S.P., Warmenhoven J.-W., Henthorn N.T., Chadiwck A.L., Santina E.E., McMahon S.J., Schuemann J., Kirkby N.F., Mackay R.I., Kirkby K.J., et al. A computational approach to quantifying miscounting of radiation-induced double-strand break immunofluorescent foci. Commun. Biol. 2022;5:700. doi: 10.1038/s42003-022-03585-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

