HPV is a cargo for the COPI sorting complex during virus entry
- PMID: 36662862
- PMCID: PMC9858521
- DOI: 10.1126/sciadv.adc9830
HPV is a cargo for the COPI sorting complex during virus entry
Abstract
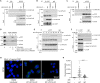
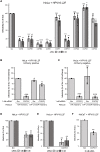
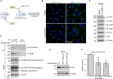
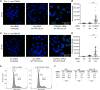
During entry, human papillomavirus (HPV) traffics from the cell surface to the endosome and then to the trans-Golgi network (TGN) and Golgi apparatus. HPV must transit across the TGN/Golgi and exit these compartments to reach the nucleus to cause infection, although how these steps are accomplished is unclear. Combining cellular fractionation, unbiased proteomics, and gene knockdown strategies, we identified the coat protein complex I (COPI), a highly conserved protein complex that facilitates retrograde trafficking of cellular cargos, as a host factor required for HPV infection. Upon TGN/Golgi arrival, the cytoplasmic segment of HPV L2 binds directly to COPI. COPI depletion causes the accumulation of HPV in the TGN/Golgi, resembling the fate of a COPI binding-defective L2 mutant. We propose that the L2-COPI interaction drives HPV trafficking through the TGN and Golgi stacks during virus entry. This shows that an incoming virus is a cargo of the COPI complex.
Figures


Similar articles
-
Rab6a enables BICD2/dynein-mediated trafficking of human papillomavirus from the trans-Golgi network during virus entry.mBio. 2024 Nov 13;15(11):e0281124. doi: 10.1128/mbio.02811-24. Epub 2024 Oct 21. mBio. 2024. PMID: 39431827 Free PMC article.
-
The BICD2 dynein cargo adaptor binds to the HPV16 L2 capsid protein and promotes HPV infection.PLoS Pathog. 2024 Jun 3;20(6):e1012289. doi: 10.1371/journal.ppat.1012289. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38829892 Free PMC article.
-
Human Papillomavirus 16 Infection Induces VAP-Dependent Endosomal Tubulation.J Virol. 2018 Feb 26;92(6):e01514-17. doi: 10.1128/JVI.01514-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29321327 Free PMC article.
-
Structure and mechanism of COPI vesicle biogenesis.Curr Opin Cell Biol. 2014 Aug;29:67-73. doi: 10.1016/j.ceb.2014.04.009. Epub 2014 May 17. Curr Opin Cell Biol. 2014. PMID: 24840894 Review.
-
Breaking the COPI monopoly on Golgi recycling.Trends Cell Biol. 2000 Sep;10(9):385-91. doi: 10.1016/s0962-8924(00)01818-3. Trends Cell Biol. 2000. PMID: 10932096 Review.
Cited by
-
Noncanonical Rab9a action supports retromer-mediated endosomal exit of human papillomavirus during virus entry.PLoS Pathog. 2023 Sep 13;19(9):e1011648. doi: 10.1371/journal.ppat.1011648. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37703297 Free PMC article.
-
Rab6a enables BICD2/dynein-mediated trafficking of human papillomavirus from the trans-Golgi network during virus entry.mBio. 2024 Nov 13;15(11):e0281124. doi: 10.1128/mbio.02811-24. Epub 2024 Oct 21. mBio. 2024. PMID: 39431827 Free PMC article.
-
Sequence-independent activity of a predicted long disordered segment of the human papillomavirus type 16 L2 capsid protein during virus entry.Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2307721120. doi: 10.1073/pnas.2307721120. Epub 2023 Oct 11. Proc Natl Acad Sci U S A. 2023. PMID: 37819982 Free PMC article.
-
The BICD2 dynein cargo adaptor binds to the HPV16 L2 capsid protein and promotes HPV infection.PLoS Pathog. 2024 Jun 3;20(6):e1012289. doi: 10.1371/journal.ppat.1012289. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38829892 Free PMC article.
-
Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.J Microbiol. 2023 Aug;61(8):755-764. doi: 10.1007/s12275-023-00071-3. Epub 2023 Sep 8. J Microbiol. 2023. PMID: 37684534
References
-
- CDC, “Cancers Associated with Human Papillomavirus, United States—2014–2018,” USCS Data Brief, no. 26 (Centers for Disease Control and Prevention, US Department of Health and Human Services, Atlanta, GA, 2021).
-
- J. C. Cardoso, E. Calonje, Cutaneous manifestations of human papillomaviruses: A review. Acta Dermatovenerol. Alp. Pannonica Adriat. 20, 145–154 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

