Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing
- PMID: 36630919
- PMCID: PMC9839364
- DOI: 10.1016/j.immuni.2022.12.004
Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing
Abstract
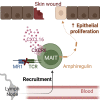
Tissue repair processes maintain proper organ function following mechanical or infection-related damage. In addition to antibacterial properties, mucosal associated invariant T (MAIT) cells express a tissue repair transcriptomic program and promote skin wound healing when expanded. Herein, we use a human-like mouse model of full-thickness skin excision to assess the underlying mechanisms of MAIT cell tissue repair function. Single-cell RNA sequencing analysis suggested that skin MAIT cells already express a repair program at steady state. Following skin excision, MAIT cells promoted keratinocyte proliferation, thereby accelerating healing. Using skin grafts, parabiosis, and adoptive transfer experiments, we show that MAIT cells migrated into the wound in a T cell receptor (TCR)-independent but CXCR6 chemokine receptor-dependent manner. Amphiregulin secreted by MAIT cells following excision promoted wound healing. Expression of the repair function was probably independent of sustained TCR stimulation. Overall, our study provides mechanistic insights into MAIT cell wound healing function in the skin.
Keywords: CXCR6 chemokine receptor; MAIT cells; TCR signaling; amphiregulin; scRNA-seq; skin excision; tissue repair.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Similar articles
-
Human liver CD8+ MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15.J Hepatol. 2020 Sep;73(3):640-650. doi: 10.1016/j.jhep.2020.03.033. Epub 2020 Apr 2. J Hepatol. 2020. PMID: 32247824
-
Mucosal-associated invariant T cells promote ductular reaction through amphiregulin in biliary atresia.EBioMedicine. 2024 May;103:105138. doi: 10.1016/j.ebiom.2024.105138. Epub 2024 Apr 27. EBioMedicine. 2024. PMID: 38678809 Free PMC article.
-
A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.J Exp Med. 2014 Jul 28;211(8):1585-600. doi: 10.1084/jem.20140484. Epub 2014 Jul 21. J Exp Med. 2014. PMID: 25049336 Free PMC article.
-
MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules?Immunogenetics. 2016 Aug;68(8):537-48. doi: 10.1007/s00251-016-0927-9. Epub 2016 Jul 8. Immunogenetics. 2016. PMID: 27393664 Review.
-
Signals that control MAIT cell function in healthy and inflamed human tissues.Immunol Rev. 2024 May;323(1):138-149. doi: 10.1111/imr.13325. Epub 2024 Mar 22. Immunol Rev. 2024. PMID: 38520075 Review.
Cited by
-
Protective effect of TCR-mediated MAIT cell activation during experimental autoimmune encephalomyelitis.Nat Commun. 2024 Oct 28;15(1):9287. doi: 10.1038/s41467-024-53657-9. Nat Commun. 2024. PMID: 39468055 Free PMC article.
-
SFRP2 modulates functional phenotype transition and energy metabolism of macrophages during diabetic wound healing.Front Immunol. 2024 Oct 11;15:1432402. doi: 10.3389/fimmu.2024.1432402. eCollection 2024. Front Immunol. 2024. PMID: 39464880 Free PMC article.
-
MAIT cells: Conserved watchers on the wall.J Exp Med. 2025 Jan 6;222(1):e20232298. doi: 10.1084/jem.20232298. Epub 2024 Oct 24. J Exp Med. 2025. PMID: 39446132 Free PMC article. Review.
-
Riboflavin for women's health and emerging microbiome strategies.NPJ Biofilms Microbiomes. 2024 Oct 18;10(1):107. doi: 10.1038/s41522-024-00579-5. NPJ Biofilms Microbiomes. 2024. PMID: 39420006 Free PMC article. Review.
-
Varicella Zoster Virus disrupts MAIT cell polyfunctional effector responses.PLoS Pathog. 2024 Aug 7;20(8):e1012372. doi: 10.1371/journal.ppat.1012372. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39110717 Free PMC article.
References
-
- Constantinides M.G., Link V.M., Tamoutounour S., Wong A.C., Perez-Chaparro P.J., Han S.-J., Chen Y.E., Li K., Farhat S., Weckel A., et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366:eaax6624. doi: 10.1126/science.aax6624. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

