The E3 Ligase TRIM25 Impairs Apoptotic Cell Death in Colon Carcinoma Cells via Destabilization of Caspase-7 mRNA: A Possible Role of hnRNPH1
- PMID: 36611995
- PMCID: PMC9818768
- DOI: 10.3390/cells12010201
The E3 Ligase TRIM25 Impairs Apoptotic Cell Death in Colon Carcinoma Cells via Destabilization of Caspase-7 mRNA: A Possible Role of hnRNPH1
Abstract
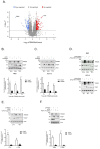
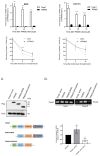
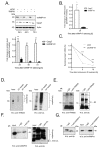
Therapy resistance is still a major reason for treatment failure in colorectal cancer (CRC). Previously, we identified the E3 ubiquitin ligase TRIM25 as a novel suppressor of caspase-2 translation which contributes to the apoptosis resistance of CRC cells towards chemotherapeutic drugs. Here, we report the executioner caspase-7 as being a further target of TRIM25. The results from the gain- and loss-of-function approaches and the actinomycin D experiments indicate that TRIM25 attenuates caspase-7 expression mainly through a decrease in mRNA stability. The data from the RNA pulldown assays with immunoprecipitated TRIM25 truncations indicate a direct TRIM25 binding to caspase-7 mRNA, which is mediated by the PRY/SPRY domain, which is also known to be highly relevant for protein-protein interactions. By employing TRIM25 immunoprecipitation, we identified the heterogeneous nuclear ribonucleoprotein H1 (hnRNPH1) as a novel TRIM25 binding protein with a functional impact on caspase-7 mRNA stability. Notably, the interaction of both proteins was highly sensitive to RNase A treatment and again depended on the PRY/SPRY domain, thus indicating an indirect interaction of both proteins which is achieved through a common RNA binding. Ubiquitin affinity chromatography showed that both proteins are targets of ubiquitin modification. Functionally, the ectopic expression of caspase-7 in CRC cells caused an increase in poly ADP-ribose polymerase (PARP) cleavage concomitant with a significant increase in apoptosis. Collectively, the negative regulation of caspase-7 by TRIM25, which is possibly executed by hnRNPH1, implies a novel survival mechanism underlying the chemotherapeutic drug resistance of CRC cells. The targeting of TRIM25 could therefore offer a promising strategy for the reduction in therapy resistance in CRC patients.
Keywords: RNA-binding proteins; TRIM25; apoptosis; caspase-7; colon carcinoma cells; hnRNPH1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Identification of TRIM25 as a Negative Regulator of Caspase-2 Expression Reveals a Novel Target for Sensitizing Colon Carcinoma Cells to Intrinsic Apoptosis.Cells. 2019 Dec 12;8(12):1622. doi: 10.3390/cells8121622. Cells. 2019. PMID: 31842382 Free PMC article.
-
RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination.BMC Biol. 2017 Nov 8;15(1):105. doi: 10.1186/s12915-017-0444-9. BMC Biol. 2017. PMID: 29117863 Free PMC article.
-
The ubiquitin ligase TRIM25 inhibits hepatocellular carcinoma progression by targeting metastasis associated 1 protein.IUBMB Life. 2017 Oct;69(10):795-801. doi: 10.1002/iub.1661. Epub 2017 Aug 31. IUBMB Life. 2017. PMID: 28861931
-
The role of Trim25 in development, disease and RNA metabolism.Biochem Soc Trans. 2016 Aug 15;44(4):1045-50. doi: 10.1042/BST20160077. Biochem Soc Trans. 2016. PMID: 27528750 Review.
-
TRIM25 and its emerging RNA-binding roles in antiviral defense.Wiley Interdiscip Rev RNA. 2020 Jul;11(4):e1588. doi: 10.1002/wrna.1588. Epub 2020 Jan 28. Wiley Interdiscip Rev RNA. 2020. PMID: 31990130 Review.
Cited by
-
The cross talk of ubiquitination and chemotherapy tolerance in colorectal cancer.J Cancer Res Clin Oncol. 2024 Mar 23;150(3):154. doi: 10.1007/s00432-024-05659-9. J Cancer Res Clin Oncol. 2024. PMID: 38521878 Free PMC article. Review.
References
-
- Papamichael D., Audisio R.A., Glimelius B., de Gramont A., Glynne-Jones R., Haller D., Köhne C.H., Rostoft S., Lemmens V., Mitry E., et al. Treatment of colorectal cancer in older patients: International Society of Geriatric Oncology (SIOG) consensus recommendations 2013. Ann. Oncol. 2015;26:463–476. doi: 10.1093/annonc/mdu253. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

