How to build a protoribosome: structural insights from the first protoribosome constructs that have proven to be catalytically active
- PMID: 36604112
- PMCID: PMC9945445
- DOI: 10.1261/rna.079417.122
How to build a protoribosome: structural insights from the first protoribosome constructs that have proven to be catalytically active
Abstract
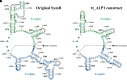
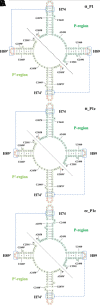
The modern ribosome catalyzes all coded protein synthesis in extant organisms. It is likely that its core structure is a direct descendant from the ribosome present in the last common ancestor (LCA). Hence, its earliest origins likely predate the LCA and therefore date further back in time. Of special interest is the pseudosymmetrical region (SymR) that lies deep within the large subunit (LSU) where the peptidyl transfer reaction takes place. It was previously proposed that two RNA oligomers, representing the P- and A-regions of extant ribosomes dimerized to create a pore-like structure, which hosted the necessary properties that facilitate peptide bond formation. However, recent experimental studies show that this may not be the case. Instead, several RNA constructs derived exclusively from the P-region were shown to form a homodimer capable of peptide bond synthesis. Of special interest will be the origin issues because the homodimer would have allowed a pre-LCA ribosome that was significantly smaller than previously proposed. For the A-region, the immediate issue will likely be its origin and whether it enhances ribosome performance. Here, we reanalyze the RNA/RNA interaction regions that most likely lead to SymR formation in light of these recent findings. Further, it has been suggested that the ability of these RNA constructs to dimerize and enhance peptide bond formation is sequence-dependent. We have analyzed the implications of sequence variations as parts of functional and nonfunctional constructs.
Keywords: RNA World; SymR; origin of life; protoribosome.
© 2023 Rivas and Fox; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
On the Re-Creation of Protoribosome Analogues in the Lab.Int J Mol Sci. 2024 May 2;25(9):4960. doi: 10.3390/ijms25094960. Int J Mol Sci. 2024. PMID: 38732179 Free PMC article. Review.
-
Origin of life: protoribosome forms peptide bonds and links RNA and protein dominated worlds.Nucleic Acids Res. 2022 Feb 28;50(4):1815-1828. doi: 10.1093/nar/gkac052. Nucleic Acids Res. 2022. PMID: 35137169 Free PMC article.
-
An exit cavity was crucial to the polymerase activity of the early ribosome.Astrobiology. 2012 Jan;12(1):57-60. doi: 10.1089/ast.2011.0692. Epub 2011 Dec 22. Astrobiology. 2012. PMID: 22191510 Free PMC article.
-
Protoribosome by quantum kernel energy method.Proc Natl Acad Sci U S A. 2013 Sep 10;110(37):14900-5. doi: 10.1073/pnas.1314112110. Epub 2013 Aug 26. Proc Natl Acad Sci U S A. 2013. PMID: 23980159 Free PMC article.
-
Origin and evolution of the ribosome.Cold Spring Harb Perspect Biol. 2010 Sep;2(9):a003483. doi: 10.1101/cshperspect.a003483. Epub 2010 Jun 9. Cold Spring Harb Perspect Biol. 2010. PMID: 20534711 Free PMC article. Review.
Cited by
-
Ancestry of RNA/RNA interaction regions within segmented ribosomes.RNA. 2023 Sep;29(9):1388-1399. doi: 10.1261/rna.079654.123. Epub 2023 Jun 1. RNA. 2023. PMID: 37263782 Free PMC article.
-
Ribosomal Protein Cluster Organization in Asgard Archaea.Archaea. 2023 Sep 29;2023:5512414. doi: 10.1155/2023/5512414. eCollection 2023. Archaea. 2023. PMID: 38314098 Free PMC article.
-
On the Re-Creation of Protoribosome Analogues in the Lab.Int J Mol Sci. 2024 May 2;25(9):4960. doi: 10.3390/ijms25094960. Int J Mol Sci. 2024. PMID: 38732179 Free PMC article. Review.
-
On the Nature of the Last Common Ancestor: A Story from its Translation Machinery.J Mol Evol. 2024 Oct;92(5):593-604. doi: 10.1007/s00239-024-10199-4. Epub 2024 Sep 11. J Mol Evol. 2024. PMID: 39259330 Review.
References
-
- Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen H, Harms J, Kessler M, et al. 2003. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Eur J Biochem 270: 2543–2556. 10.1046/j.1432-1033.2003.03634.x - DOI - PubMed
-
- Agmon I, Davidovich C, Bashan A, Yonath A. 2009. Identification of the prebiotic translation apparatus within the contemporary ribosome. Nat Preced 10.1038/npre.2009.2921.1 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
