Human Endogenous Retrovirus (HERV) Transcriptome Is Dynamically Modulated during SARS-CoV-2 Infection and Allows Discrimination of COVID-19 Clinical Stages
- PMID: 36602345
- PMCID: PMC9927238
- DOI: 10.1128/spectrum.02516-22
Human Endogenous Retrovirus (HERV) Transcriptome Is Dynamically Modulated during SARS-CoV-2 Infection and Allows Discrimination of COVID-19 Clinical Stages
Abstract
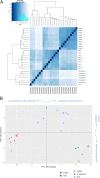
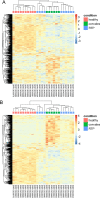
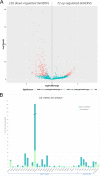
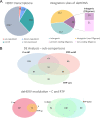
SARS-CoV-2 infection is known to trigger an important inflammatory response, which has a major role in COVID-19 pathogenesis. In infectious and inflammatory contexts, the modulation of human endogenous retroviruses (HERV) has been broadly reported, being able to further sustain innate immune responses due to the expression of immunogenic viral transcripts, including double-stranded DNA (dsRNA), and eventually, immunogenic proteins. To gain insights on this poorly characterized interplay, we performed a high-throughput expression analysis of ~3,300 specific HERV loci in the peripheral blood mononuclear cells (PBMCs) of 10 healthy controls and 16 individuals being either convalescent after the infection (6) or retesting positive after convalescence (10) (Gene Expression Onmibus [GEO] data set GSE166253). Results showed that the exposure to SARS-CoV-2 infection modulates HERV expression according to the disease stage and reflecting COVID-19 immune signatures. The differential expression analysis between healthy control (HC) and COVID-19 patients allowed us to identify a total of 282 differentially expressed HERV loci (deHERV) in the individuals exposed to SARS-CoV-2 infection, independently from the clinical form. In addition, 278 and 60 deHERV loci that were specifically modulated in individuals convalescent after COVID19 infection (C) and patients that retested positive to SARS-CoV-2 after convalescence (RTP) as individually compared to HC, respectively, as well as 164 deHERV loci between C and RTP patients were identified. The identified HERV loci belonged to 36 different HERV groups, including members of all three classes. The present study provides an exhaustive picture of the HERV transcriptome in PBMCs and its dynamic variation in the presence of COVID-19, revealing specific modulation patterns according to the infection stage that can be relevant to the disease clinical manifestation and outcome. IMPORTANCE We report here the first high-throughput analysis of HERV loci expression along SARS-CoV-2 infection, as performed with peripheral blood mononuclear cells (PBMCs). Such cells are not directly infected by the virus but have a crucial role in the plethora of inflammatory and immune events that constitute a major hallmark of COVID-19 pathogenesis. Results provide a novel and exhaustive picture of HERV expression in PBMCs, revealing specific modulation patterns according to the disease condition and the concomitant immune activation. To our knowledge, this is the first set of deHERVs whose expression is dynamically modulated across COVID-19 stages, confirming a tight interplay between HERV and cellular immunity and revealing specific transcriptional signatures that can have an impact on the disease clinical manifestation and outcome.
Keywords: COVID-19; HERV; RNA-seq; SARS-CoV-2; human endogenous retroviruses; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Shared and unique patterns of autonomous human endogenous retrovirus loci transcriptomes in CD14 + monocytes from individuals with physical trauma or infection with COVID-19.Retrovirology. 2024 Nov 4;21(1):17. doi: 10.1186/s12977-024-00652-z. Retrovirology. 2024. PMID: 39497142 Free PMC article.
-
Upregulation of Human Endogenous Retroviruses in Bronchoalveolar Lavage Fluid of COVID-19 Patients.Microbiol Spectr. 2021 Oct 31;9(2):e0126021. doi: 10.1128/Spectrum.01260-21. Epub 2021 Oct 6. Microbiol Spectr. 2021. PMID: 34612698 Free PMC article.
-
RNA-Seq Transcriptome Analysis Reveals Long Terminal Repeat Retrotransposon Modulation in Human Peripheral Blood Mononuclear Cells after In Vivo Lipopolysaccharide Injection.J Virol. 2020 Sep 15;94(19):e00587-20. doi: 10.1128/JVI.00587-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669333 Free PMC article.
-
Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses.Front Immunol. 2018 Sep 10;9:2039. doi: 10.3389/fimmu.2018.02039. eCollection 2018. Front Immunol. 2018. PMID: 30250470 Free PMC article. Review.
-
Type W Human Endogenous Retrovirus (HERV-W) Integrations and Their Mobilization by L1 Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology.Viruses. 2017 Jun 27;9(7):162. doi: 10.3390/v9070162. Viruses. 2017. PMID: 28653997 Free PMC article. Review.
Cited by
-
Human endogenous retroviruses and exogenous viral infections.Front Cell Infect Microbiol. 2024 Sep 27;14:1439292. doi: 10.3389/fcimb.2024.1439292. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39397863 Free PMC article. Review.
-
Long COVID: plasma levels of neurofilament light chain in mild COVID-19 patients with neurocognitive symptoms.Mol Psychiatry. 2024 Oct;29(10):3106-3116. doi: 10.1038/s41380-024-02554-0. Epub 2024 Apr 27. Mol Psychiatry. 2024. PMID: 38678084 Free PMC article.
-
HERVs: Expression Control Mechanisms and Interactions in Diseases and Human Immunodeficiency Virus Infection.Genes (Basel). 2024 Jan 31;15(2):192. doi: 10.3390/genes15020192. Genes (Basel). 2024. PMID: 38397182 Free PMC article. Review.
-
Exploring HERV-K (HML-2) Influence in Cancer and Prospects for Therapeutic Interventions.Int J Mol Sci. 2023 Sep 27;24(19):14631. doi: 10.3390/ijms241914631. Int J Mol Sci. 2023. PMID: 37834078 Free PMC article. Review.
-
Navigating Evolving Challenges in Blood Safety.Viruses. 2024 Jan 15;16(1):123. doi: 10.3390/v16010123. Viruses. 2024. PMID: 38257823 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

