Werner syndrome protein works as a dimer for unwinding and replication fork regression
- PMID: 36583333
- PMCID: PMC9841404
- DOI: 10.1093/nar/gkac1200
Werner syndrome protein works as a dimer for unwinding and replication fork regression
Abstract
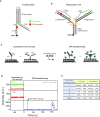
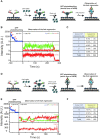
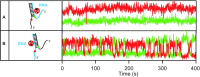
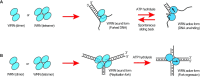
The determination of the oligomeric state of functional enzymes is essential for the mechanistic understanding of their catalytic activities. RecQ helicases have diverse biochemical activities, but it is still unclear how their activities are related to their oligomeric states. We use single-molecule multi-color fluorescence imaging to determine the oligomeric states of Werner syndrome protein (WRN) during its unwinding and replication fork regression activities. We reveal that WRN binds to a forked DNA as a dimer, and unwinds it without any change of its oligomeric state. In contrast, WRN binds to a replication fork as a tetramer, and is dimerized during activation of replication fork regression. By selectively inhibiting the helicase activity of WRN on specific strands, we reveal how the active dimers of WRN distinctly use the energy of ATP hydrolysis for repetitive unwinding and replication fork regression.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity.Nucleic Acids Res. 2007;35(17):5729-47. doi: 10.1093/nar/gkm561. Epub 2007 Aug 23. Nucleic Acids Res. 2007. PMID: 17717003 Free PMC article.
-
The Werner and Bloom syndrome proteins catalyze regression of a model replication fork.Biochemistry. 2006 Nov 28;45(47):13939-46. doi: 10.1021/bi0615487. Biochemistry. 2006. PMID: 17115688
-
Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage.J Biol Chem. 2011 Feb 4;286(5):3497-508. doi: 10.1074/jbc.M110.105411. Epub 2010 Nov 24. J Biol Chem. 2011. PMID: 21107010 Free PMC article.
-
The Werner syndrome protein: linking the replication checkpoint response to genome stability.Aging (Albany NY). 2011 Mar;3(3):311-8. doi: 10.18632/aging.100293. Aging (Albany NY). 2011. PMID: 21389352 Free PMC article. Review.
-
Roles of the Werner syndrome RecQ helicase in DNA replication.DNA Repair (Amst). 2008 Nov 1;7(11):1776-86. doi: 10.1016/j.dnarep.2008.07.017. Epub 2008 Sep 6. DNA Repair (Amst). 2008. PMID: 18722555 Free PMC article. Review.
Cited by
-
Repair and tolerance of DNA damage at the replication fork: A structural perspective.Curr Opin Struct Biol. 2023 Aug;81:102618. doi: 10.1016/j.sbi.2023.102618. Epub 2023 Jun 1. Curr Opin Struct Biol. 2023. PMID: 37269798 Free PMC article. Review.
-
Alteration of replication protein A binding mode on single-stranded DNA by NSMF potentiates RPA phosphorylation by ATR kinase.Nucleic Acids Res. 2023 Aug 25;51(15):7936-7950. doi: 10.1093/nar/gkad543. Nucleic Acids Res. 2023. PMID: 37378431 Free PMC article.
References
-
- Ellis N.A., Groden J., Ye T.-Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J.. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995; 83:655–666. - PubMed
-
- Yu C.-E., Oshima J., Fu Y.-H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S.. Positional cloning of the Werner's syndrome gene. Science. 1996; 272:258–262. - PubMed
-
- Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y.. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999; 22:82–84. - PubMed
-
- Siitonen H.A., Kopra O., Kääriäinen H., Haravuori H., Winter R.M., Säämänen A.-M., Peltonen L., Kestilä M.. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003; 12:2837–2844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

