Truvari: refined structural variant comparison preserves allelic diversity
- PMID: 36575487
- PMCID: PMC9793516
- DOI: 10.1186/s13059-022-02840-6
Truvari: refined structural variant comparison preserves allelic diversity
Abstract
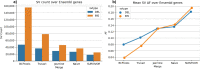
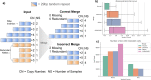
The fundamental challenge of multi-sample structural variant (SV) analysis such as merging and benchmarking is identifying when two SVs are the same. Common approaches for comparing SVs were developed alongside technologies which produce ill-defined boundaries. As SV detection becomes more exact, algorithms to preserve this refined signal are needed. Here, we present Truvari-an SV comparison, annotation, and analysis toolkit-and demonstrate the effect of SV comparison choices by building population-level VCFs from 36 haplotype-resolved long-read assemblies. We observe over-merging from other SV merging approaches which cause up to a 2.2× inflation of allele frequency, relative to Truvari.
Keywords: SV annotation; SV benchmarking; SV comparison; SV merging; Structural variation.
© 2022. The Author(s).
Conflict of interest statement
FJS received research support from PacBio and Oxford Nanopore.
Figures


Similar articles
-
VISTA: an integrated framework for structural variant discovery.Brief Bioinform. 2024 Jul 25;25(5):bbae462. doi: 10.1093/bib/bbae462. Brief Bioinform. 2024. PMID: 39297879 Free PMC article.
-
svclassify: a method to establish benchmark structural variant calls.BMC Genomics. 2016 Jan 16;17:64. doi: 10.1186/s12864-016-2366-2. BMC Genomics. 2016. PMID: 26772178 Free PMC article.
-
Robust Benchmark Structural Variant Calls of An Asian Using State-of-the-art Long-read Sequencing Technologies.Genomics Proteomics Bioinformatics. 2022 Feb;20(1):192-204. doi: 10.1016/j.gpb.2020.10.006. Epub 2021 Mar 2. Genomics Proteomics Bioinformatics. 2022. PMID: 33662625 Free PMC article.
-
Cataloging Plant Genome Structural Variations.Curr Issues Mol Biol. 2018;27:181-194. doi: 10.21775/cimb.027.181. Epub 2017 Sep 8. Curr Issues Mol Biol. 2018. PMID: 28885182 Review.
-
Applications of advanced technologies for detecting genomic structural variation.Mutat Res Rev Mutat Res. 2023 Jul-Dec;792:108475. doi: 10.1016/j.mrrev.2023.108475. Epub 2023 Nov 4. Mutat Res Rev Mutat Res. 2023. PMID: 37931775 Free PMC article. Review.
Cited by
-
Nanopore sequencing of 1000 Genomes Project samples to build a comprehensive catalog of human genetic variation.medRxiv [Preprint]. 2024 Mar 7:2024.03.05.24303792. doi: 10.1101/2024.03.05.24303792. medRxiv. 2024. Update in: Genome Res. 2024 Nov 20;34(11):2061-2073. doi: 10.1101/gr.279273.124 PMID: 38496498 Free PMC article. Updated. Preprint.
-
AsmMix: an efficient haplotype-resolved hybrid de novo genome assembling pipeline.Front Genet. 2024 Jul 26;15:1421565. doi: 10.3389/fgene.2024.1421565. eCollection 2024. Front Genet. 2024. PMID: 39130747 Free PMC article.
-
INSnet: a method for detecting insertions based on deep learning network.BMC Bioinformatics. 2023 Mar 6;24(1):80. doi: 10.1186/s12859-023-05216-0. BMC Bioinformatics. 2023. PMID: 36879189 Free PMC article.
-
Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation.Nat Methods. 2023 Oct;20(10):1483-1492. doi: 10.1038/s41592-023-01993-x. Epub 2023 Sep 14. Nat Methods. 2023. PMID: 37710018 Free PMC article.
-
Comprehensive de novo mutation discovery with HiFi long-read sequencing.Genome Med. 2023 May 8;15(1):34. doi: 10.1186/s13073-023-01183-6. Genome Med. 2023. PMID: 37158973 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

