Diallyl Disulfide Induces Chemosensitization to Sorafenib, Autophagy, and Cell Cycle Arrest and Inhibits Invasion in Hepatocellular Carcinoma
- PMID: 36559076
- PMCID: PMC9788602
- DOI: 10.3390/pharmaceutics14122582
Diallyl Disulfide Induces Chemosensitization to Sorafenib, Autophagy, and Cell Cycle Arrest and Inhibits Invasion in Hepatocellular Carcinoma
Abstract
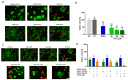
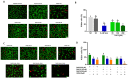
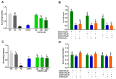
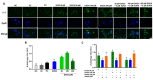
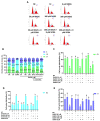
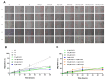
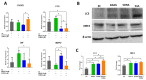
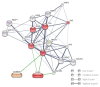
Hepatocellular carcinoma is the seventh most common type of cancer in the world, with limited treatment options. A promising strategy to treat cancer is to associate chemotherapeutics and plant bioactive compounds. Here, we examined whether diallyl disulfide (DADS; 50-200 μM) and sorafenib (SORA; 8 μM), either alone or in combination, were toxic to hepatocellular carcinoma cells (HepG2) in vitro. We assessed whether DADS and/or SORA induced cell death (LIVE/DEAD assay and autophagy) and cell cycle changes (flow cytometry), altered expression of key genes and proteins (RT-qPCR and Western blot), and modulated tumorigenesis signatures, such as proliferation (clonogenic assay), migration (wound healing), and invasion (inserts). The DADS + SORA combination elicited autophagic cell death by upregulating LC3 and NRF2 expression and downregulating FOS and TNF expression; induced the accumulation of cells in the G1 phase which thereby upregulated the CHEK2 expression; and inhibited invasion by downregulating the MMP2 expression. Predictive analysis indicated the participation of the MAPK pathway in the reported results. The DADS + SORA combination suppressed both cell invasion and clonogenic survival, which indicated that it dampened tumor growth, proliferation, invasion, and metastatic potential. Therefore, the DADS + SORA combination is a promising therapy to develop new clinical protocols.
Keywords: Allium sativum L.; DNA damage; liver cancer; nutraceutical; nutrigenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Pristimerin synergistically sensitizes conditionally reprogrammed patient derived-primary hepatocellular carcinoma cells to sorafenib through endoplasmic reticulum stress and ROS generation by modulating Akt/FoxO1/p27kip1 signaling pathway.Phytomedicine. 2021 Jun;86:153563. doi: 10.1016/j.phymed.2021.153563. Epub 2021 Apr 4. Phytomedicine. 2021. PMID: 33951569
-
HANR Enhances Autophagy-Associated Sorafenib Resistance Through miR-29b/ATG9A Axis in Hepatocellular Carcinoma.Onco Targets Ther. 2020 Mar 9;13:2127-2137. doi: 10.2147/OTT.S229913. eCollection 2020. Onco Targets Ther. 2020. PMID: 32210579 Free PMC article.
-
Diallyl Disulfide Induces Apoptosis and Autophagy in Human Osteosarcoma MG-63 Cells through the PI3K/Akt/mTOR Pathway.Molecules. 2019 Jul 23;24(14):2665. doi: 10.3390/molecules24142665. Molecules. 2019. PMID: 31340526 Free PMC article.
-
Diallyl sulfide, diallyl disulfide, and diallyl trisulfide inhibit migration and invasion in human colon cancer colo 205 cells through the inhibition of matrix metalloproteinase-2, -7, and -9 expressions.Environ Toxicol. 2013 Sep;28(9):479-88. doi: 10.1002/tox.20737. Epub 2011 Jun 21. Environ Toxicol. 2013. PMID: 21695758
-
Uvangoletin, extracted from Sarcandra glabra, exerts anticancer activity by inducing autophagy and apoptosis and inhibiting invasion and migration on hepatocellular carcinoma cells.Phytomedicine. 2022 Jan;94:153793. doi: 10.1016/j.phymed.2021.153793. Epub 2021 Oct 4. Phytomedicine. 2022. PMID: 34736000
Cited by
-
D-arabinose induces cell cycle arrest by promoting autophagy via p38 MAPK signaling pathway in breast cancer.Sci Rep. 2024 May 16;14(1):11219. doi: 10.1038/s41598-024-61309-7. Sci Rep. 2024. PMID: 38755221 Free PMC article.
-
Pectinose induces cell cycle arrest in luminal A and triple-negative breast cancer cells by promoting autophagy through activation of the p38 MAPK signaling pathway.BMC Cancer. 2024 May 24;24(1):639. doi: 10.1186/s12885-024-12293-8. BMC Cancer. 2024. PMID: 38789954 Free PMC article.
-
Sulforaphane Combined with Vitamin D Induces Cytotoxicity Mediated by Oxidative Stress, DNA Damage, Autophagy, and JNK/MAPK Pathway Modulation in Human Prostate Tumor Cells.Nutrients. 2023 Jun 14;15(12):2742. doi: 10.3390/nu15122742. Nutrients. 2023. PMID: 37375646 Free PMC article.
-
Diallyl disulfide synergizes with melphalan to increase apoptosis and DNA damage through elevation of reactive oxygen species in multiple myeloma cells.Ann Hematol. 2024 Apr;103(4):1293-1303. doi: 10.1007/s00277-023-05592-w. Epub 2023 Dec 27. Ann Hematol. 2024. PMID: 38148345
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

