RNA Epigenetics in Chronic Lung Diseases
- PMID: 36553648
- PMCID: PMC9777603
- DOI: 10.3390/genes13122381
RNA Epigenetics in Chronic Lung Diseases
Abstract
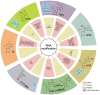
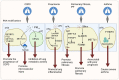
Chronic lung diseases are highly prevalent worldwide and cause significant mortality. Lung cancer is the end stage of many chronic lung diseases. RNA epigenetics can dynamically modulate gene expression and decide cell fate. Recently, studies have confirmed that RNA epigenetics plays a crucial role in the developing of chronic lung diseases. Further exploration of the underlying mechanisms of RNA epigenetics in chronic lung diseases, including lung cancer, may lead to a better understanding of the diseases and promote the development of new biomarkers and therapeutic strategies. This article reviews basic information on RNA modifications, including N6 methylation of adenosine (m6A), N1 methylation of adenosine (m1A), N7-methylguanosine (m7G), 5-methylcytosine (m5C), 2'O-methylation (2'-O-Me or Nm), pseudouridine (5-ribosyl uracil or Ψ), and adenosine to inosine RNA editing (A-to-I editing). We then show how they relate to different types of lung disease. This paper hopes to summarize the mechanisms of RNA modification in chronic lung disease and finds a new way to develop early diagnosis and treatment of chronic lung disease.
Keywords: RNA epigenetics; RNA methylation; chronic lung disease; lung cancer; treatment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases.Int J Mol Sci. 2021 Sep 30;22(19):10592. doi: 10.3390/ijms221910592. Int J Mol Sci. 2021. PMID: 34638933 Free PMC article. Review.
-
RNA epigenetics and cardiovascular diseases.J Mol Cell Cardiol. 2019 Apr;129:272-280. doi: 10.1016/j.yjmcc.2019.03.010. Epub 2019 Mar 14. J Mol Cell Cardiol. 2019. PMID: 30880252 Free PMC article. Review.
-
RNA modifications in cardiovascular diseases, the potential therapeutic targets.Life Sci. 2021 Aug 1;278:119565. doi: 10.1016/j.lfs.2021.119565. Epub 2021 May 6. Life Sci. 2021. PMID: 33965380 Review.
-
RNA modifications: importance in immune cell biology and related diseases.Signal Transduct Target Ther. 2022 Sep 22;7(1):334. doi: 10.1038/s41392-022-01175-9. Signal Transduct Target Ther. 2022. PMID: 36138023 Free PMC article. Review.
-
The effects of RNA methylation on immune cells development and function.FASEB J. 2022 Oct;36(10):e22552. doi: 10.1096/fj.202200716R. FASEB J. 2022. PMID: 36111987 Review.
Cited by
-
Epigenetic regulation of pulmonary inflammation.Semin Cell Dev Biol. 2024 Feb 15;154(Pt C):346-354. doi: 10.1016/j.semcdb.2023.05.003. Epub 2023 May 23. Semin Cell Dev Biol. 2024. PMID: 37230854 Review.
-
Identification and validation of key biomarkers based on RNA methylation genes in sepsis.Front Immunol. 2023 Aug 28;14:1231898. doi: 10.3389/fimmu.2023.1231898. eCollection 2023. Front Immunol. 2023. PMID: 37701433 Free PMC article.
-
Inflammation during Pregnancy Predisposes to Childhood Asthma by Altering Long Noncoding RNA Expression.Am J Respir Cell Mol Biol. 2023 Jun;68(6):599-600. doi: 10.1165/rcmb.2023-0004ED. Am J Respir Cell Mol Biol. 2023. PMID: 36701649 Free PMC article. No abstract available.
-
RNA modifications in pulmonary diseases.MedComm (2020). 2024 May 3;5(5):e546. doi: 10.1002/mco2.546. eCollection 2024 May. MedComm (2020). 2024. PMID: 38706740 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

