ON-1 and BA-IX Are the Dominant Sub-Genotypes of Human Orthopneumovirus A&B in Riyadh, Saudi Arabia
- PMID: 36553555
- PMCID: PMC9778264
- DOI: 10.3390/genes13122288
ON-1 and BA-IX Are the Dominant Sub-Genotypes of Human Orthopneumovirus A&B in Riyadh, Saudi Arabia
Abstract
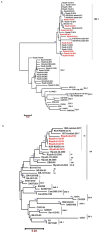
Human orthopneumovirus (HOPV) is the major viral pathogen responsible for lower respiratory tract infections (LRTIs) in infants and young children in Riyadh, Saudi Arabia. Yet, predominant HOPV subtypes circulating in this region and their molecular and epidemiological characteristics are not fully ascertained. A total of 300 clinical samples involving nasopharyngeal aspirates (NPAs), throat swabs, and sputum were collected during winter seasons of 2019/2020 and 2021/2022 for HOPV subtyping and genotyping. Of the 300 samples, HOPV was identified in 55 samples (18.3%) with a distinct predominance of type A viruses (81.8%) compared to type B viruses (18.2%). Importantly, the ON1 strain of HOPV-A and BA-IX strain of HOPV-B groups were found to be responsible for all the infections. Sequence analysis revealed a duplication region within 2nd HVR of G protein gene of ON1 and BA-IX strains. This nucleotide duplication exerted a profound effect on protein length and affinity towards cell receptors. Further, these modifications may aid the HOPV in immune evasion and recurrent infections. Data from this study showed that ON-1 genotype of HOPV-A and BA-IX genotype of HOPV-B were dominant in Riyadh, Saudi Arabia. Further, a duplication of sequence within 2nd HVR of G protein gene was found.
Keywords: 2nd HVR; BA-IX; HRSV; ON-1; Riyadh; Saudi Arabia; human orthopneumovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Co-Circulation of 72bp Duplication Group A and 60bp Duplication Group B Respiratory Syncytial Virus (RSV) Strains in Riyadh, Saudi Arabia during 2014.PLoS One. 2016 Nov 11;11(11):e0166145. doi: 10.1371/journal.pone.0166145. eCollection 2016. PLoS One. 2016. PMID: 27835664 Free PMC article.
-
Evolutionary analysis of the ON1 genotype of subtype a respiratory syncytial virus in Riyadh during 2008-16.Infect Genet Evol. 2020 Apr;79:104153. doi: 10.1016/j.meegid.2019.104153. Epub 2019 Dec 24. Infect Genet Evol. 2020. PMID: 31881360
-
The emergence of subgenotype ON-1 of Human orthopneumovirus type A in Riyadh, Saudi Arabia: A new episode of the virus epidemiological dynamic.J Med Virol. 2020 Aug;92(8):1133-1140. doi: 10.1002/jmv.25643. Epub 2019 Dec 9. J Med Virol. 2020. PMID: 31777964
-
Genetic diversity in the G protein gene of group A human respiratory syncytial viruses circulating in Riyadh, Saudi Arabia.Arch Virol. 2014 Jan;159(1):73-81. doi: 10.1007/s00705-013-1792-6. Epub 2013 Jul 25. Arch Virol. 2014. PMID: 23884633
-
Molecular Characterization of Human Respiratory Syncytial Virus in the Philippines, 2012-2013.PLoS One. 2015 Nov 5;10(11):e0142192. doi: 10.1371/journal.pone.0142192. eCollection 2015. PLoS One. 2015. PMID: 26540236 Free PMC article.
References
-
- Mazur N.I., Martinón-Torres F., Baraldi E., Fauroux B., Greenough A., Heikkinen T., Manzoni P., Mejias A., Nair H., Papadopoulos N.G., et al. Lower respiratory tract infection caused by respiratory syncytial virus: Current management and new therapeutics. Lancet Respir. Med. 2015;3:888–900. doi: 10.1016/S2213-2600(15)00255-6. - DOI - PubMed
-
- Mazur N.I., Bont L., Cohen A.L., Cohen C., Von Gottberg A., Groome M.J., Hellferscee O., Klipstein-Grobusch K., Mekgoe O.T., Naby F., et al. Severity of Respiratory Syncytial Virus Lower Respiratory Tract Infection with Viral Coinfection in HIV-Uninfected Children. Clin. Infect. Dis. 2017;64:443–450. doi: 10.1093/cid/ciw756. - DOI - PMC - PubMed
-
- Zhong Q., Feng H., Lü Q., Liu X., Zhao Q., Du Y., Zhang X.-H., Wang J.-R. Recurrent wheezing in neonatal pneumonia is associated with combined infection with Respiratory Syncytial Virus and Staphylococcus aureus or Klebsiella pneumoniae. Sci. Rep. 2018;8:995. doi: 10.1038/s41598-018-19386-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

