The deubiquitinase USP10 protects pancreatic cancer cells from endoplasmic reticulum stress
- PMID: 36543867
- PMCID: PMC9772324
- DOI: 10.1038/s41698-022-00336-x
The deubiquitinase USP10 protects pancreatic cancer cells from endoplasmic reticulum stress
Abstract
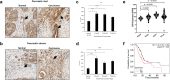
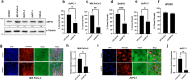
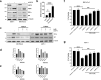
The ubiquitin-specific peptidase 10 (USP10) plays a context-specific, pro or anti-tumorigenic role in different malignancies. However, the role of USP10 in pancreatic cancer remains unclear. Our protein and RNA level analysis from archived specimens and public databases show that USP10 is overexpressed in pancreatic ductal adenocarcinoma (PDAC) and expression correlates with poor overall patient survival. Phenotypically, silencing USP10 decreased viability, clonal growth and invasive properties of pancreatic cancer cells. Mechanistically, silencing USP10 upregulated BiP and induced endoplasmic reticulum (ER) stress that led to an unfolded protein response (UPR) and upregulation of PERK, IRE1α. Decreased cell viability of USP10 silenced cells could be rescued by a chemical chaperone that promotes protein folding. Our studies suggest that USP10 by protecting pancreatic cancer cells from ER stress may support tumor progression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Inhibition of endoplasmic-reticulum-stress-mediated autophagy enhances the effectiveness of chemotherapeutics on pancreatic cancer.J Transl Med. 2018 Jul 9;16(1):190. doi: 10.1186/s12967-018-1562-z. J Transl Med. 2018. PMID: 29986726 Free PMC article.
-
Deubiquitinating PABPC1 by USP10 upregulates CLK2 translation to promote tumor progression in pancreatic ductal adenocarcinoma.Cancer Lett. 2023 Nov 1;576:216411. doi: 10.1016/j.canlet.2023.216411. Epub 2023 Sep 26. Cancer Lett. 2023. PMID: 37757903
-
Deubiquitinase ubiquitin-specific peptidase 10 maintains cysteine rich angiogenic inducer 61 expression via Yes1 associated transcriptional regulator to augment immune escape and metastasis of pancreatic adenocarcinoma.Cancer Sci. 2022 May;113(5):1868-1879. doi: 10.1111/cas.15326. Epub 2022 Apr 5. Cancer Sci. 2022. PMID: 35271750 Free PMC article.
-
The unfolded protein response: An emerging therapeutic target for pancreatitis and pancreatic ductal adenocarcinoma.Pancreatology. 2022 Jan;22(1):148-159. doi: 10.1016/j.pan.2021.10.007. Epub 2021 Oct 28. Pancreatology. 2022. PMID: 34774415 Review.
-
Dual role of Endoplasmic Reticulum Stress-Mediated Unfolded Protein Response Signaling Pathway in Carcinogenesis.Int J Mol Sci. 2019 Sep 5;20(18):4354. doi: 10.3390/ijms20184354. Int J Mol Sci. 2019. PMID: 31491919 Free PMC article. Review.
Cited by
-
USP10 promotes pancreatic ductal adenocarcinoma progression by attenuating FOXC1 protein degradation to activate the WNT signaling pathway.Int J Biol Sci. 2024 Sep 30;20(13):5343-5362. doi: 10.7150/ijbs.92278. eCollection 2024. Int J Biol Sci. 2024. PMID: 39430239 Free PMC article.
-
USP10 promotes intrahepatic cholangiocarcinoma cell survival and stemness via SNAI1 deubiquitination.J Mol Histol. 2023 Dec;54(6):703-714. doi: 10.1007/s10735-023-10150-9. Epub 2023 Sep 27. J Mol Histol. 2023. PMID: 37755617
-
Stress granules in cancer: Adaptive dynamics and therapeutic implications.iScience. 2024 Jun 22;27(8):110359. doi: 10.1016/j.isci.2024.110359. eCollection 2024 Aug 16. iScience. 2024. PMID: 39100690 Free PMC article. Review.
-
The role of deubiquitinases in cardiac disease.Expert Rev Mol Med. 2024 Mar 25;26:e3. doi: 10.1017/erm.2024.2. Expert Rev Mol Med. 2024. PMID: 38525836 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

