Insights into divalent cation regulation and G13-coupling of orphan receptor GPR35
- PMID: 36543774
- PMCID: PMC9772185
- DOI: 10.1038/s41421-022-00499-8
Insights into divalent cation regulation and G13-coupling of orphan receptor GPR35
Abstract
Endogenous ions play important roles in the function and pharmacology of G protein-coupled receptors (GPCRs) with limited atomic evidence. In addition, compared with G protein subtypes Gs, Gi/o, and Gq/11, insufficient structural evidence is accessible to understand the coupling mechanism of G12/13 protein by GPCRs. Orphan receptor GPR35, which is predominantly expressed in the gastrointestinal tract and is closely related to inflammatory bowel diseases (IBDs), stands out as a prototypical receptor for investigating ionic modulation and G13 coupling. Here we report a cryo-electron microscopy structure of G13-coupled GPR35 bound to an anti-allergic drug, lodoxamide. This structure reveals a novel divalent cation coordination site and a unique ionic regulatory mode of GPR35 and also presents a highly positively charged binding pocket and the complementary electrostatic ligand recognition mode, which explain the promiscuity of acidic ligand binding by GPR35. Structural comparison of the GPR35-G13 complex with other G protein subtypes-coupled GPCRs reveals a notable movement of the C-terminus of α5 helix of the Gα13 subunit towards the receptor core and the least outward displacement of the cytoplasmic end of GPR35 TM6. A featured 'methionine pocket' contributes to the G13 coupling by GPR35. Together, our findings provide a structural basis for divalent cation modulation, ligand recognition, and subsequent G13 protein coupling of GPR35 and offer a new opportunity for designing GPR35-targeted drugs for the treatment of IBDs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
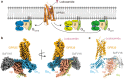



Similar articles
-
Receptor selectivity between the G proteins Gα12 and Gα13 is defined by a single leucine-to-isoleucine variation.FASEB J. 2019 Apr;33(4):5005-5017. doi: 10.1096/fj.201801956R. Epub 2019 Jan 2. FASEB J. 2019. PMID: 30601679 Free PMC article.
-
Isoforms of GPR35 have distinct extracellular N-termini that allosterically modify receptor-transducer coupling and mediate intracellular pathway bias.J Biol Chem. 2022 Sep;298(9):102328. doi: 10.1016/j.jbc.2022.102328. Epub 2022 Aug 4. J Biol Chem. 2022. PMID: 35933013 Free PMC article.
-
Receptor-dependent RhoA activation in G12/G13-deficient cells: genetic evidence for an involvement of Gq/G11.J Biol Chem. 2003 Aug 1;278(31):28743-9. doi: 10.1074/jbc.M304570200. Epub 2003 May 27. J Biol Chem. 2003. PMID: 12771155
-
The therapeutic potential of orphan GPCRs, GPR35 and GPR55.Front Pharmacol. 2015 Apr 15;6:69. doi: 10.3389/fphar.2015.00069. eCollection 2015. Front Pharmacol. 2015. PMID: 25926795 Free PMC article. Review.
-
The emerging pharmacology and function of GPR35 in the nervous system.Neuropharmacology. 2017 Feb;113(Pt B):661-671. doi: 10.1016/j.neuropharm.2015.07.035. Epub 2015 Jul 29. Neuropharmacology. 2017. PMID: 26232640 Review.
Cited by
-
Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice.Mar Drugs. 2024 Apr 15;22(4):175. doi: 10.3390/md22040175. Mar Drugs. 2024. PMID: 38667792 Free PMC article.
-
Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.Cell. 2024 Aug 22;187(17):4751-4769.e25. doi: 10.1016/j.cell.2024.07.005. Epub 2024 Jul 31. Cell. 2024. PMID: 39089252 Free PMC article.
-
Mast cell stabilizers: from pathogenic roles to targeting therapies.Front Immunol. 2024 Aug 1;15:1418897. doi: 10.3389/fimmu.2024.1418897. eCollection 2024. Front Immunol. 2024. PMID: 39148726 Free PMC article. Review.
-
GPR35 and mediators from platelets and mast cells in neutrophil migration and inflammation.Immunol Rev. 2023 Aug;317(1):187-202. doi: 10.1111/imr.13194. Epub 2023 Mar 16. Immunol Rev. 2023. PMID: 36928841 Free PMC article. Review.
-
GPR35 acts a dual role and therapeutic target in inflammation.Front Immunol. 2023 Nov 16;14:1254446. doi: 10.3389/fimmu.2023.1254446. eCollection 2023. Front Immunol. 2023. PMID: 38035084 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

