Thermodynamic analysis of an entropically driven, high-affinity nanobody-HIV p24 interaction
- PMID: 36527237
- PMCID: PMC9892613
- DOI: 10.1016/j.bpj.2022.12.019
Thermodynamic analysis of an entropically driven, high-affinity nanobody-HIV p24 interaction
Abstract
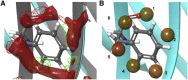
Protein-protein interactions are fundamental to life processes. Complementary computational, structural, and biophysical studies of these interactions enable the forces behind their specificity and strength to be understood. Antibody fragments such as single-chain antibodies have the specificity and affinity of full antibodies but a fraction of their size, expediting whole molecule studies and distal effects without exceeding the computational capacity of modeling systems. We previously reported the crystal structure of a high-affinity nanobody 59H10 bound to HIV-1 capsid protein p24 and deduced key interactions using all-atom molecular dynamics simulations. We studied the properties of closely related medium (37E7) and low (48G11) affinity nanobodies, to understand how changes of three (37E7) or one (48G11) amino acids impacted these interactions; however, the contributions of enthalpy and entropy were not quantified. Here, we report the use of qualitative and quantitative experimental and in silico approaches to separate the contributions of enthalpy and entropy. We used complementary circular dichroism spectroscopy and molecular dynamics simulations to qualitatively delineate changes between nanobodies in isolation and complexed with p24. Using quantitative techniques such as isothermal titration calorimetry alongside WaterMap and Free Energy Perturbation protocols, we found the difference between high (59H10) and medium (37E7) affinity nanobodies on binding to HIV-1 p24 is entropically driven, accounted for by the release of unstable waters from the hydrophobic surface of 59H10. Our results provide an exemplar of the utility of parallel in vitro and in silico studies and highlight that differences in entropic interactions between amino acids and water molecules are sufficient to drive orders of magnitude differences in affinity.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
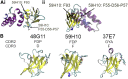

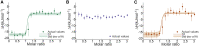
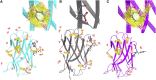
Similar articles
-
Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.ACS Infect Dis. 2017 Jul 14;3(7):479-491. doi: 10.1021/acsinfecdis.6b00189. Epub 2017 Jun 26. ACS Infect Dis. 2017. PMID: 28591513
-
Thermodynamics of Nanobody Binding to Lactose Permease.Biochemistry. 2016 Oct 25;55(42):5917-5926. doi: 10.1021/acs.biochem.6b00826. Epub 2016 Oct 12. Biochemistry. 2016. PMID: 27686537 Free PMC article.
-
Interaction of epitope-related and -unrelated peptides with anti-p24 (HIV-1) monoclonal antibody CB4-1 and its Fab fragment.J Mol Recognit. 2003 Jan-Feb;16(1):54-62. doi: 10.1002/jmr.607. J Mol Recognit. 2003. PMID: 12557239
-
Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering.Int J Mol Sci. 2022 Mar 8;23(6):2928. doi: 10.3390/ijms23062928. Int J Mol Sci. 2022. PMID: 35328351 Free PMC article. Review.
-
Direct measurement of protein binding energetics by isothermal titration calorimetry.Curr Opin Struct Biol. 2001 Oct;11(5):560-6. doi: 10.1016/s0959-440x(00)00248-7. Curr Opin Struct Biol. 2001. PMID: 11785756 Review.
References
-
- Steeland S., Vandenbroucke R.E., Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov. Today. 2016;21:1076–1113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

