Downregulation of HbFPS1 affects rubber biosynthesis of Hevea brasiliensis suffering from tapping panel dryness
- PMID: 36524729
- PMCID: PMC10107253
- DOI: 10.1111/tpj.16063
Downregulation of HbFPS1 affects rubber biosynthesis of Hevea brasiliensis suffering from tapping panel dryness
Abstract
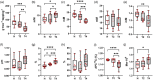
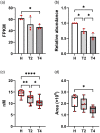
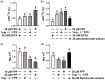
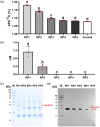
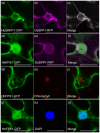
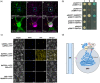
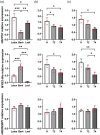
Tapping panel dryness (TPD) is a century-old problem that has plagued the natural rubber production of Hevea brasiliensis. TPD may result from self-protective mechanisms of H. brasiliensis in response to stresses such as excessive hormone stimulation and mechanical wounding (bark tapping). It has been hypothesized that TPD impairs rubber biosynthesis; however, the underlying mechanisms remain poorly understood. In the present study, we firstly verified that TPD-affected rubber trees exhibited lower rubber biosynthesis activity and greater rubber molecular weight compared to healthy rubber trees. We then demonstrated that HbFPS1, a key gene of rubber biosynthesis, and its expression products were downregulated in the latex of TPD-affected rubber trees, as revealed by transcriptome sequencing and iTRAQ-based proteome analysis. We further discovered that the farnesyl diphosphate synthase HbFPS1 could be recruited to small rubber particles by HbSRPP1 through protein-protein interactions to catalyze farnesyl diphosphate (FPP) synthesis and facilitate rubber biosynthesis initiation. FPP content in the latex of TPD-affected rubber trees was significantly decreased with the downregulation of HbFPS1, ultimately resulting in abnormal development of rubber particles, decreased rubber biosynthesis activity, and increased rubber molecular weight. Upstream regulator assays indicated that a novel regulator, MYB2-like, may be an important regulator of downregulation of HbFPS1 in the latex of TPD-affected rubber trees. Our findings not only provide new directions for studying the molecular events involved in rubber biosynthesis and TPD syndrome and contribute to rubber management strategies, but also broaden our knowledge of plant isoprenoid metabolism and its regulatory networks.
Keywords: Hevea brasiliensis; MYB transcription factor; farnesyl diphosphate synthase; iTRAQ; protein-protein interaction; rubber biosynthesis; tapping panel dryness; transcriptome sequencing.
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Bark transcriptome analyses reveals molecular mechanisms involved in tapping panel dryness occurrence and development in rubber tree (Hevea brasiliensis).Gene. 2024 Jan 20;892:147894. doi: 10.1016/j.gene.2023.147894. Epub 2023 Oct 11. Gene. 2024. PMID: 37832804
-
Transcriptome analysis in Hevea brasiliensis latex revealed changes in hormone signalling pathways during ethephon stimulation and consequent Tapping Panel Dryness.Sci Rep. 2018 May 31;8(1):8483. doi: 10.1038/s41598-018-26854-y. Sci Rep. 2018. PMID: 29855601 Free PMC article.
-
Molecular identification and characterization of a gene associated with the onset of tapping panel dryness (TPD) syndrome in rubber tree (Hevea brasiliensis Muell.) by mRNA differential display.Mol Biotechnol. 2009 Jan;41(1):42-52. doi: 10.1007/s12033-008-9095-y. Epub 2008 Aug 23. Mol Biotechnol. 2009. PMID: 18726169
-
Genomic technologies for Hevea breeding.Adv Genet. 2019;104:1-73. doi: 10.1016/bs.adgen.2019.04.001. Epub 2019 Jun 3. Adv Genet. 2019. PMID: 31200808 Review.
-
Molecular Mechanisms of Natural Rubber Biosynthesis.Annu Rev Biochem. 2020 Jun 20;89:821-851. doi: 10.1146/annurev-biochem-013118-111107. Epub 2020 Mar 30. Annu Rev Biochem. 2020. PMID: 32228045 Review.
Cited by
-
Advancing image segmentation with DBO-Otsu: Addressing rubber tree diseases through enhanced threshold techniques.PLoS One. 2024 Mar 21;19(3):e0297284. doi: 10.1371/journal.pone.0297284. eCollection 2024. PLoS One. 2024. PMID: 38512907 Free PMC article.
References
-
- Adiwilage, K. & Kush, A. (1996) Cloning and characterization of cDNA encoding farnesyl diphosphate synthase from rubber tree (Hevea brasiliensis). Plant Molecular Biology, 30, 935–946. - PubMed
-
- Chen, S.C. , Peng, S.Q. , Huang, G.X. , Wu, K.X. , Fu, X.H. & Chen, Z.Q. (2003) Association of decreased expression of a Myb transcription factor with the TPD (tapping panel drynss) syndrome in Hevea brasiliensis . Plant Molecular Biology, 51, 51–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

