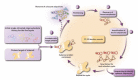
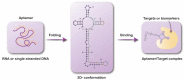
Therapeutic Potential of Aptamer-Protein Interactions
- PMID: 36524009
- PMCID: PMC9745894
- DOI: 10.1021/acsptsci.2c00156
Therapeutic Potential of Aptamer-Protein Interactions
Abstract
Aptamers are single-stranded oligonucleotides (RNA or DNA) with a typical length between 25 and 100 nucleotides which fold into three-dimensional structures capable of binding to target molecules. Specific aptamers can be isolated against a large variety of targets through efficient and relatively cheap methods, and they demonstrate target-binding affinities that sometimes surpass those of antibodies. Consequently, interest in aptamers has surged over the past three decades, and their application has shown promise in advancing knowledge in target analysis, designing therapeutic interventions, and bioengineering. With emphasis on their therapeutic applications, aptamers are emerging as a new innovative class of therapeutic agents with promising biochemical and biological properties. Aptamers have the potential of providing a feasible alternative to antibody- and small-molecule-based therapeutics given their binding specificity, stability, low toxicity, and apparent non-immunogenicity. This Review examines the general properties of aptamers and aptamer-protein interactions that help to understand their binding characteristics and make them important therapeutic candidates.
© 2022 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Investigations on the interface of nucleic acid aptamers and binding targets.Analyst. 2018 Nov 5;143(22):5317-5338. doi: 10.1039/c8an01467a. Analyst. 2018. PMID: 30357118 Review.
-
Characterisation of aptamers for therapeutic studies.Expert Opin Drug Discov. 2007 Sep;2(9):1205-24. doi: 10.1517/17460441.2.9.1205. Expert Opin Drug Discov. 2007. PMID: 23496129
-
Oligonucleotide aptamers: new tools for targeted cancer therapy.Mol Ther Nucleic Acids. 2014 Aug 5;3(8):e182. doi: 10.1038/mtna.2014.32. Mol Ther Nucleic Acids. 2014. PMID: 25093706 Free PMC article.
-
The development and testing of aptamers for cancer.Curr Opin Investig Drugs. 2009 Jun;10(6):572-8. Curr Opin Investig Drugs. 2009. PMID: 19513946 Review.
-
Aptamer-Based Targeted Drug Delivery Systems: Current Potential and Challenges.Curr Med Chem. 2020;27(13):2189-2219. doi: 10.2174/0929867325666181008142831. Curr Med Chem. 2020. PMID: 30295183 Review.
Cited by
-
Aptamers: precision tools for diagnosing and treating infectious diseases.Front Cell Infect Microbiol. 2024 Sep 25;14:1402932. doi: 10.3389/fcimb.2024.1402932. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39386170 Free PMC article. Review.
-
Unlocking precision in aptamer engineering: a case study of the thrombin binding aptamer illustrates why modification size, quantity, and position matter.Nucleic Acids Res. 2024 Oct 14;52(18):10823-10835. doi: 10.1093/nar/gkae729. Nucleic Acids Res. 2024. PMID: 39217472 Free PMC article.
-
Recent advances in surface plasmon resonance for the detection of ovarian cancer biomarkers: a thorough review.Mikrochim Acta. 2024 Oct 9;191(11):659. doi: 10.1007/s00604-024-06740-3. Mikrochim Acta. 2024. PMID: 39382786 Review.
-
AptaTrans: a deep neural network for predicting aptamer-protein interaction using pretrained encoders.BMC Bioinformatics. 2023 Nov 27;24(1):447. doi: 10.1186/s12859-023-05577-6. BMC Bioinformatics. 2023. PMID: 38012571 Free PMC article.
-
Selection of a novel cell-internalizing RNA aptamer specific for CD22 antigen in B cell acute lymphoblastic leukemia.Mol Ther Nucleic Acids. 2023 Jul 28;33:698-712. doi: 10.1016/j.omtn.2023.07.028. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37662970 Free PMC article.
References
-
- Gilbert W. Origin of life: The RNA world. Nature 1986, 319, 618.10.1038/319618a0. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous