Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta gene as a tumour suppressor in stomach adenocarcinoma
- PMID: 36518312
- PMCID: PMC9743170
- DOI: 10.3389/fonc.2022.1069875
Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta gene as a tumour suppressor in stomach adenocarcinoma
Abstract
Background: Stomach adenocarcinoma (STAD) is the most common type of gastric cancer. In this study, the functions and potential mechanisms of hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta (HADHB) in STAD were explored.
Methods: Different bioinformatics analyses were performed to confirm HADHB expression in STAD. HADHB expression in STAD tissues and cells was also evaluated using western blot, qRT-PCR, and immunohistochemistry. Further, the viability, proliferation, colony formation, cell cycle determination, migration, and wound healing capacity were assessed, and the effects of HADHB on tumour growth, cell apoptosis, and proliferation in nude mice were determined. The upstream effector of HADHB was examined using bioinformatics analysis and dual luciferase reporter assay. GSEA was also employed for pathway enrichment analysis and the expression of Hippo-YAP pathway-related proteins was detected.
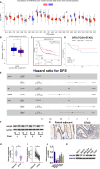
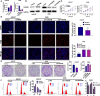
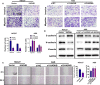
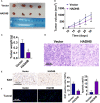
Results: The expression of HADHB was found to be low in STAD tissues and cells. The upregulation of HADHB distinctly repressed the viability, proliferation, colony formation, cell cycle progression, migration, invasion, and wound healing of HGC27 cells, while knockdown of HADHB led to opposite effects. HADHB upregulation impeded tumour growth and cell proliferation, and enhanced apoptosis in nude mice. KLF4, whose expression was low in STAD, was identified as an upstream regulator of HADHB. KLF4 upregulation abolished the HADHB knockdown-induced tumour promoting effects in AGS cells. Further, HADHB regulates the Hippo-YAP pathway, which was validated using a pathway rescue assay. Low expression of KLF4 led to HADHB downregulation in STAD.
Conclusion: HADHB might function as a tumour suppressor gene in STAD by regulation the Hippo-YAP pathway.
Keywords: HADHB; Hippo-YAP pathway; KLF4; cell proliferation; stomach adenocarcinoma.
Copyright © 2022 Li, Xiong, Jie and Xiong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
DNA demethylase TET2-mediated reduction of HADHB expression contributes to cadmium-induced malignant progression of colorectal cancer.Ecotoxicol Environ Saf. 2024 Jul 15;280:116579. doi: 10.1016/j.ecoenv.2024.116579. Epub 2024 Jun 11. Ecotoxicol Environ Saf. 2024. PMID: 38865940
-
OASL knockdown inhibits the progression of stomach adenocarcinoma by regulating the mTORC1 signaling pathway.FASEB J. 2023 Mar;37(3):e22824. doi: 10.1096/fj.202201582R. FASEB J. 2023. PMID: 36809539
-
HADHB, HuR, and CP1 bind to the distal 3'-untranslated region of human renin mRNA and differentially modulate renin expression.J Biol Chem. 2003 Nov 7;278(45):44894-903. doi: 10.1074/jbc.M307782200. Epub 2003 Aug 21. J Biol Chem. 2003. PMID: 12933794
-
TEAD4 Activates PCSK9 to Promote Stomach Adenocarcinoma Cell Stemness through Fatty Acid Metabolism.Digestion. 2024;105(4):243-256. doi: 10.1159/000538329. Epub 2024 Apr 26. Digestion. 2024. PMID: 38663369
-
HADHB, a fatty acid beta-oxidation enzyme, is a potential prognostic predictor in malignant lymphoma.Pathology. 2022 Apr;54(3):286-293. doi: 10.1016/j.pathol.2021.06.119. Epub 2021 Sep 14. Pathology. 2022. PMID: 34531036
Cited by
-
Genetics of enzymatic dysfunctions in metabolic disorders and cancer.Front Oncol. 2023 Aug 2;13:1230934. doi: 10.3389/fonc.2023.1230934. eCollection 2023. Front Oncol. 2023. PMID: 37601653 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

