Transcriptomic and splicing changes underlying tomato responses to combined water and nutrient stress
- PMID: 36507383
- PMCID: PMC9732681
- DOI: 10.3389/fpls.2022.974048
Transcriptomic and splicing changes underlying tomato responses to combined water and nutrient stress
Abstract
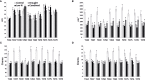
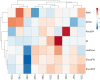
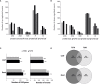
Tomato is a horticultural crop of high economic and nutritional value. Suboptimal environmental conditions, such as limited water and nutrient availability, cause severe yield reductions. Thus, selection of genotypes requiring lower inputs is a goal for the tomato breeding sector. We screened 10 tomato varieties exposed to water deficit, low nitrate or a combination of both. Biometric, physiological and molecular analyses revealed different stress responses among genotypes, identifying T270 as severely affected, and T250 as tolerant to the stresses applied. Investigation of transcriptome changes caused by combined stress in roots and leaves of these two genotypes yielded a low number of differentially expressed genes (DEGs) in T250 compared to T270, suggesting that T250 tailors changes in gene expression to efficiently respond to combined stress. By contrast, the susceptible tomato activated approximately one thousand and two thousand genes in leaves and roots respectively, indicating a more generalized stress response in this genotype. In particular, developmental and stress-related genes were differentially expressed, such as hormone responsive factors and transcription factors. Analysis of differential alternative splicing (DAS) events showed that combined stress greatly affects the splicing landscape in both genotypes, highlighting the important role of AS in stress response mechanisms. In particular, several stress and growth-related genes as well as transcription and splicing factors were differentially spliced in both tissues. Taken together, these results reveal important insights into the transcriptional and post-transcriptional mechanisms regulating tomato adaptation to growth under reduced water and nitrogen inputs.
Keywords: differential alternative splicing; differential gene expression; leaf; low nitrate; root; tolerant and sensitive genotypes; water deficit.
Copyright © 2022 Ruggiero, Punzo, Van Oosten, Cirillo, Esposito, Costa, Maggio, Grillo and Batelli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
RNA-seq analysis reveals transcriptome reprogramming and alternative splicing during early response to salt stress in tomato root.Front Plant Sci. 2024 Jun 20;15:1394223. doi: 10.3389/fpls.2024.1394223. eCollection 2024. Front Plant Sci. 2024. PMID: 38966147 Free PMC article.
-
Genome-wide transcriptional analysis of two soybean genotypes under dehydration and rehydration conditions.BMC Genomics. 2013 Oct 6;14:687. doi: 10.1186/1471-2164-14-687. BMC Genomics. 2013. PMID: 24093224 Free PMC article.
-
Comparative transcriptomic and physiological analyses of contrasting hybrid cultivars ND476 and ZX978 identify important differentially expressed genes and pathways regulating drought stress tolerance in maize.Genes Genomics. 2020 Aug;42(8):937-955. doi: 10.1007/s13258-020-00962-4. Epub 2020 Jul 4. Genes Genomics. 2020. PMID: 32623576
-
Root transcriptome profiling of contrasting wheat genotypes provides an insight to their adaptive strategies to water deficit.Sci Rep. 2020 Mar 17;10(1):4854. doi: 10.1038/s41598-020-61680-1. Sci Rep. 2020. PMID: 32184417 Free PMC article.
-
Root-Related Genes in Crops and Their Application under Drought Stress Resistance-A Review.Int J Mol Sci. 2022 Sep 29;23(19):11477. doi: 10.3390/ijms231911477. Int J Mol Sci. 2022. PMID: 36232779 Free PMC article. Review.
Cited by
-
Nonsense-mediated mRNA decay pathway in plants under stress: general gene regulatory mechanism and advances.Planta. 2024 Jan 30;259(3):51. doi: 10.1007/s00425-023-04317-7. Planta. 2024. PMID: 38289504 Review.
-
Alternative Splicing Variation: Accessing and Exploiting in Crop Improvement Programs.Int J Mol Sci. 2023 Oct 15;24(20):15205. doi: 10.3390/ijms242015205. Int J Mol Sci. 2023. PMID: 37894886 Free PMC article. Review.
-
Young Tomato Plants Respond Differently under Single or Combined Mild Nitrogen and Water Deficit: An Insight into Morphophysiological Responses and Primary Metabolism.Plants (Basel). 2023 Mar 5;12(5):1181. doi: 10.3390/plants12051181. Plants (Basel). 2023. PMID: 36904041 Free PMC article.
-
RNA-seq analysis reveals transcriptome reprogramming and alternative splicing during early response to salt stress in tomato root.Front Plant Sci. 2024 Jun 20;15:1394223. doi: 10.3389/fpls.2024.1394223. eCollection 2024. Front Plant Sci. 2024. PMID: 38966147 Free PMC article.
References
-
- Bouzroud S., Gouiaa S., Hu N., Bernadac A., Mila I., Bendaou N., et al. . (2018). Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PloS One 13 (2), e0193517. doi: 10.1371/journal.pone.0193517 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

