The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction
- PMID: 36506089
- PMCID: PMC9732275
- DOI: 10.3389/fcell.2022.1053139
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction
Abstract
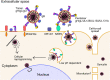
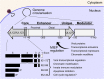
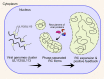
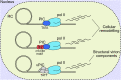
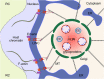
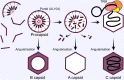
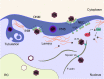
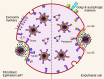
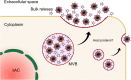
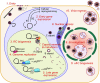
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen that can cause severe disease in immunocompromised individuals, transplant recipients, and to the developing foetus during pregnancy. There is no protective vaccine currently available, and with only a limited number of antiviral drug options, resistant strains are constantly emerging. Successful completion of HCMV replication is an elegant feat from a molecular perspective, with both host and viral processes required at various stages. Remarkably, HCMV and other herpesviruses have protracted replication cycles, large genomes, complex virion structure and complicated nuclear and cytoplasmic replication events. In this review, we outline the 10 essential stages the virus must navigate to successfully complete replication. As each individual event along the replication continuum poses as a potential barrier for restriction, these essential checkpoints represent potential targets for antiviral development.
Keywords: HCMV (human cytomegalovirus); antiviral therapeutic; herpes viral infection; viral replication; virion assembly.
Copyright © 2022 Turner and Mathias.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage.J Virol. 2014 Jun;88(12):6970-82. doi: 10.1128/JVI.00384-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696486 Free PMC article.
-
Asparagine Deprivation Causes a Reversible Inhibition of Human Cytomegalovirus Acute Virus Replication.mBio. 2019 Oct 8;10(5):e01651-19. doi: 10.1128/mBio.01651-19. mBio. 2019. PMID: 31594813 Free PMC article.
-
Identification of Host Factors Involved in Human Cytomegalovirus Replication, Assembly, and Egress Using a Two-Step Small Interfering RNA Screen.mBio. 2018 Jun 26;9(3):e00716-18. doi: 10.1128/mBio.00716-18. mBio. 2018. PMID: 29946045 Free PMC article.
-
Drug targets in cytomegalovirus infection.Infect Disord Drug Targets. 2009 Apr;9(2):201-22. doi: 10.2174/187152609787847758. Infect Disord Drug Targets. 2009. PMID: 19275707 Review.
-
Early inhibitors of human cytomegalovirus: state-of-art and therapeutic perspectives.Pharmacol Ther. 2011 Sep;131(3):309-29. doi: 10.1016/j.pharmthera.2011.04.007. Epub 2011 Apr 28. Pharmacol Ther. 2011. PMID: 21570424 Free PMC article. Review.
Cited by
-
Understanding the Cytomegalovirus Cyclin-Dependent Kinase Ortholog pUL97 as a Multifaceted Regulator and an Antiviral Drug Target.Cells. 2024 Aug 13;13(16):1338. doi: 10.3390/cells13161338. Cells. 2024. PMID: 39195228 Free PMC article. Review.
-
Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism.J Virol. 2023 Aug 31;97(8):e0078123. doi: 10.1128/jvi.00781-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565748 Free PMC article.
-
Secondary Envelopment of Human Cytomegalovirus Is a Fast Process Utilizing the Endocytic Compartment as a Major Membrane Source.Biomolecules. 2024 Sep 12;14(9):1149. doi: 10.3390/biom14091149. Biomolecules. 2024. PMID: 39334915 Free PMC article.
-
Cytomegaloviruses reorganize endomembrane system to intersect endosomal and amphisome-like egress pathway.Front Cell Dev Biol. 2023 Dec 19;11:1328751. doi: 10.3389/fcell.2023.1328751. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38178873 Free PMC article. No abstract available.
-
'Getting Better'-Is It a Feasible Strategy of Broad Pan-Antiherpesviral Drug Targeting by Using the Nuclear Egress-Directed Mechanism?Int J Mol Sci. 2024 Feb 29;25(5):2823. doi: 10.3390/ijms25052823. Int J Mol Sci. 2024. PMID: 38474070 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

