Identification of metabolism-related genes for predicting peritoneal metastasis in patients with gastric cancer
- PMID: 36503378
- PMCID: PMC9743729
- DOI: 10.1186/s12863-022-01096-0
Identification of metabolism-related genes for predicting peritoneal metastasis in patients with gastric cancer
Abstract
Objective: The reprogramming of metabolism is an important factor in the metastatic process of cancer. In our study, we intended to investigate the predictive value of metabolism-related genes (MRGs) in recurrent gastric cancer (GC) patients with peritoneal metastasis.
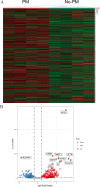
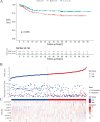
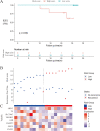
Methods: The sequencing data of mRNA of GC patients were obtained from Asian Cancer Research Group (ACRG) and the GEO databases (GSE53276). The differentially expressed MRGs (DE-MRGs) between a cell line without peritoneal metastasis (HSC60) and one with peritoneal metastasis (60As6) were analyzed with the Limma package. According to the LASSO regression, eight MRGs were identified as crucially related to peritoneal seeding recurrence in patients. Then, disease free survival related genes were screened using Cox regression, and a promising prognostic model was constructed based on 8 MRGs. We trained and verified it in two independent cohort.
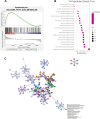
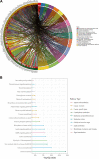
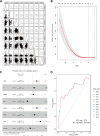
Results: We confirmed 713 DE-MRGs and the enriched pathways. Pathway analysis found that the MRG-related pathways were related to tumor metabolism development. With the help of Kaplan-Meier analysis, we found that the group with higher risk scores had worse rates of peritoneal seeding recurrence than the group with lower scores in the cohorts.
Conclusions: This study developed an eight-gene signature correlated with metabolism that could predict peritoneal seeding recurrence for GC patients. This signature could be a promising prognostic model, providing better strategy in treatment.
Keywords: Gastric cancer; Gene; Metabolism; Peritoneal metastasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Immune Landscape of Gastric Carcinoma Tumor Microenvironment Identifies a Peritoneal Relapse Relevant Immune Signature.Front Immunol. 2021 May 13;12:651033. doi: 10.3389/fimmu.2021.651033. eCollection 2021. Front Immunol. 2021. PMID: 34054812 Free PMC article.
-
Identification of the angiogenesis related genes for predicting prognosis of patients with gastric cancer.BMC Gastroenterol. 2021 Apr 1;21(1):146. doi: 10.1186/s12876-021-01734-4. BMC Gastroenterol. 2021. PMID: 33794777 Free PMC article.
-
A novel signature derived from metabolism-related genes GPT and SMS to predict prognosis of laryngeal squamous cell carcinoma.Cancer Cell Int. 2022 Jul 8;22(1):226. doi: 10.1186/s12935-022-02647-2. Cancer Cell Int. 2022. PMID: 35804447 Free PMC article.
-
Identification of a metabolism-related gene expression prognostic model in endometrial carcinoma patients.BMC Cancer. 2020 Sep 7;20(1):864. doi: 10.1186/s12885-020-07345-8. BMC Cancer. 2020. PMID: 32894095 Free PMC article.
-
Prognostic Value and Correlation With Tumor Immune Infiltration of a Novel Metabolism-Related Gene Signature in Pancreatic Cancer.Front Oncol. 2022 Jan 19;11:757791. doi: 10.3389/fonc.2021.757791. eCollection 2021. Front Oncol. 2022. PMID: 35127473 Free PMC article.
Cited by
-
Development of a Signature Based on Eight Metastatic-Related Genes for Prognosis of GC Patients.Mol Biotechnol. 2023 Nov;65(11):1796-1808. doi: 10.1007/s12033-023-00671-9. Epub 2023 Feb 15. Mol Biotechnol. 2023. PMID: 36790659 Free PMC article.
-
Discovery of CDK signature and CDK5 as potential biomarkers for predicting prognosis and immunotherapeutic response in gastric cancer peritoneal metastases.Am J Cancer Res. 2023 Sep 15;13(9):4087-4100. eCollection 2023. Am J Cancer Res. 2023. PMID: 37818084 Free PMC article.
-
Gastric cancer peritoneal metastasis related signature predicts prognosis and sensitivity to immunotherapy in gastric cancer.J Cell Mol Med. 2023 Nov;27(22):3578-3590. doi: 10.1111/jcmm.17922. Epub 2023 Aug 21. J Cell Mol Med. 2023. PMID: 37605453 Free PMC article.
-
Elucidation of population stratifying markers and selective sweeps in crossbred Landlly pig population using genome-wide SNP data.Mamm Genome. 2024 Jun;35(2):170-185. doi: 10.1007/s00335-024-10029-4. Epub 2024 Mar 15. Mamm Genome. 2024. PMID: 38485788
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
