Consequences of somatic mutations of GIRK1 detected in primary malign tumors on expression and function of G-protein activated, inwardly rectifying, K+ channels
- PMID: 36483038
- PMCID: PMC9724741
- DOI: 10.3389/fonc.2022.998907
Consequences of somatic mutations of GIRK1 detected in primary malign tumors on expression and function of G-protein activated, inwardly rectifying, K+ channels
Abstract
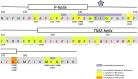
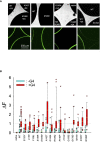
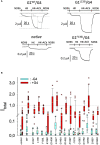
A search in the GDC Data Portal revealed 304 documented somatic mutations of the KCNJ3 gene in primary tumors (out of 10.202 cases). Most affected tumor types were carcinomas from uterus, skin and lung, while breast cancer exerted the lowest number of somatic mutations. We focused our research on 15 missense mutations within the region between TM1 and TM2, comprising the pore helix and ion selectivity signature. Expression was measured by confocal laser scan microscopy of eGFP tagged GIRK1 subunits, expressed with and without GIRK4 in oocytes of Xenopus laevis. GIRK ion currents were activated via coexpressed m2Rs and measured by the Two Electrode Voltage Clamp technique. Magnitude of the total GIRK current, as well as the fraction of current inducible by the agonist, were measured. Ion selectivity was gauged by assessment of the PNa+/PK+ ratio, calculated by the GIRK current reversal potential in extracellular media at different Na+ and K+ concentrations. None of the tested mutations was able to form functional GIRK1 homooligomeric ion channels. One of the mutations, G145A, which locates directly to the ion selectivity signature, exerted an increased PNa+/PK+ ratio. Generally, the missense mutations studied can be categorized into three groups: (i) normal/reduced expression accompanied by reduced/absent function (S132Y, F136L, E139K, G145A, R149Q, R149P, G178D, S185Y, Q186R), (ii) normal/increased expression as well as increased function (E140M, A142T, M184I) and (iii) miniscule expression but increased function relative to expression levels (I151N, G158S). We conclude, that gain of function mutations, identical or similar to categories (ii) and (iii), may potentially be involved in genesis and progression of malignancies in tissues that exert a high rate of occurrence of somatic mutations of KCNJ3.
Keywords: GIRK1; Xenopus laevis (X. laevis); confocal laser scanning microcopy; ion channels and cancer; somatic mutation analysis; two electrode voltage clamp.
Copyright © 2022 Pelzmann, Hatab, Scheruebel, Langthaler, Rienmueller, Sokolowski, Gorischek, Platzer, Zorn-Pauly, Jahn, Bauernhofer and Schreibmayer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A revised mechanism of action of hyperaldosteronism-linked mutations in cytosolic domains of GIRK4 (KCNJ5).J Physiol. 2022 Mar;600(6):1419-1437. doi: 10.1113/JP282690. Epub 2022 Jan 17. J Physiol. 2022. PMID: 34957562
-
Asymmetrical contributions of subunit pore regions to ion selectivity in an inward rectifier K+ channel.Biophys J. 1998 Sep;75(3):1330-9. doi: 10.1016/S0006-3495(98)74051-2. Biophys J. 1998. PMID: 9726934 Free PMC article.
-
Heterologous facilitation of G protein-activated K(+) channels by beta-adrenergic stimulation via cAMP-dependent protein kinase.J Gen Physiol. 2000 May;115(5):547-58. doi: 10.1085/jgp.115.5.547. J Gen Physiol. 2000. PMID: 10779313 Free PMC article.
-
G protein-gated inwardly rectifying potassium channels are targets for volatile anesthetics.Mol Pharmacol. 2001 Aug;60(2):282-9. doi: 10.1124/mol.60.2.282. Mol Pharmacol. 2001. PMID: 11455015
-
G-protein mediated gating of inward-rectifier K+ channels.Eur J Biochem. 2000 Oct;267(19):5830-6. doi: 10.1046/j.1432-1327.2000.01670.x. Eur J Biochem. 2000. PMID: 10998041 Review.
References
LinkOut - more resources
Full Text Sources

