Regulation of ribonucleoprotein condensates by RNase L during viral infection
- PMID: 36479619
- PMCID: PMC10244490
- DOI: 10.1002/wrna.1770
Regulation of ribonucleoprotein condensates by RNase L during viral infection
Abstract
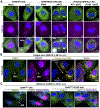
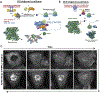
In response to viral infection, mammalian cells activate several innate immune pathways to antagonize viral gene expression. Upon recognition of viral double-stranded RNA, protein kinase R (PKR) phosphorylates the alpha subunit of eukaryotic initiation factor 2 (eIF2α) on serine 51. This inhibits canonical translation initiation, which broadly antagonizes viral protein synthesis. It also promotes the assembly of cytoplasmic ribonucleoprotein complexes termed stress granules (SGs). SGs are widely thought to promote cell survival and antiviral signaling. However, co-activation of the OAS/RNase L antiviral pathway inhibits the assembly of SGs and promotes the assembly of an alternative ribonucleoprotein complex termed an RNase L-dependent body (RLB). The formation of RLBs has been observed in response to double-stranded RNA, dengue virus infection, or SARS-CoV-2 infection. Herein, we review the distinct biogenesis pathways and properties of SGs and RLBs, and we provide perspective on their potential functions during the antiviral response. This article is categorized under: RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes RNA Turnover and Surveillance > Regulation of RNA Stability RNA Export and Localization > RNA Localization.
Keywords: RNase L; RNase L-dependent body; innate immunity; stress granules; virus.
© 2022 The Author. WIREs RNA published by Wiley Periodicals LLC.
Conflict of interest statement
Figures
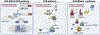


Similar articles
-
A closer look at mammalian antiviral condensates.Biochem Soc Trans. 2024 Jun 26;52(3):1393-1404. doi: 10.1042/BST20231296. Biochem Soc Trans. 2024. PMID: 38778761 Free PMC article. Review.
-
RNase L promotes the formation of unique ribonucleoprotein granules distinct from stress granules.J Biol Chem. 2020 Feb 7;295(6):1426-1438. doi: 10.1074/jbc.RA119.011638. Epub 2020 Jan 2. J Biol Chem. 2020. PMID: 31896577 Free PMC article.
-
The Nucleocapsid Proteins of SARS-CoV-2 and Its Close Relative Bat Coronavirus RaTG13 Are Capable of Inhibiting PKR- and RNase L-Mediated Antiviral Pathways.Microbiol Spectr. 2023 Jun 15;11(3):e0099423. doi: 10.1128/spectrum.00994-23. Epub 2023 May 8. Microbiol Spectr. 2023. PMID: 37154717 Free PMC article.
-
SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes.Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2022643118. doi: 10.1073/pnas.2022643118. Proc Natl Acad Sci U S A. 2021. PMID: 33811184 Free PMC article.
-
RNA regulation of the antiviral protein 2'-5'-oligoadenylate synthetase.Wiley Interdiscip Rev RNA. 2019 Jul;10(4):e1534. doi: 10.1002/wrna.1534. Epub 2019 Apr 15. Wiley Interdiscip Rev RNA. 2019. PMID: 30989826 Free PMC article. Review.
Cited by
-
RNase L-induced bodies sequester subgenomic flavivirus RNAs and re-establish host RNA decay.bioRxiv [Preprint]. 2024 Mar 27:2024.03.25.586660. doi: 10.1101/2024.03.25.586660. bioRxiv. 2024. Update in: Cell Rep. 2024 Sep 24;43(9):114694. doi: 10.1016/j.celrep.2024.114694 PMID: 38585896 Free PMC article. Updated. Preprint.
-
A closer look at mammalian antiviral condensates.Biochem Soc Trans. 2024 Jun 26;52(3):1393-1404. doi: 10.1042/BST20231296. Biochem Soc Trans. 2024. PMID: 38778761 Free PMC article. Review.
References
-
- Brennan-Laun SE, Li XL, Ezelle HJ, Venkataraman T, Blackshear PJ, Wilson GM, & Hassel BA (2014). RNase L attenuates mitogen-stimulated gene expression via transcriptional and post-transcriptional mechanisms to limit the proliferative response. The Journal of biological chemistry, 289(48), 33629–33643. 10.1074/jbc.M114.589556 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

