Genetic dissection of the soybean dwarf mutant dm with integrated genomic, transcriptomic and methylomic analyses
- PMID: 36479521
- PMCID: PMC9721362
- DOI: 10.3389/fpls.2022.1017672
Genetic dissection of the soybean dwarf mutant dm with integrated genomic, transcriptomic and methylomic analyses
Abstract
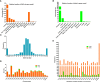
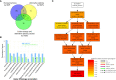
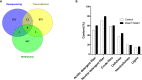
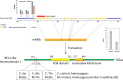
Plant height affects crop production and breeding practices, while genetic control of dwarfism draws a broad interest of researchers. Dwarfism in soybean (Glycine max) is mainly unexplored. Here, we characterized a dwarf mutant dm screened from ethyl methanesulfonate (EMS) mutated seeds of the soybean cultivar Zhongpin 661(ZP). Phenotypically, dm showed shorter and thinner stems, smaller leaves, and more nodes than ZP under greenhouse conditions. Genetically, whole-genome sequencing and comparison revealed that 210K variants of SNPs and InDel in ZP relative to the soybean reference genome Williams82, and EMS mutagenesis affected 636 genes with variants predicted to have a large impact on protein function in dm. Whole-genome methylation sequencing found 704 differentially methylated regions in dm. Further whole-genome RNA-Seq based transcriptomic comparison between ZP and dm leaves revealed 687 differentially expressed genes (DEGs), including 263 up-regulated and 424 down-regulated genes. Integrated omics analyses revealed 11 genes with both differential expressions and DNA variants, one gene with differential expression and differential methylation, and three genes with differential methylation and sequence variation, worthy of future investigation. Genes in cellulose, fatty acids, and energy-associated processes could be the key candidate genes for the dwarf phenotype. This study provides genetic clues for further understanding of the genetic control of dwarfism in soybean. The genetic resources could help to inbreed new cultivars with a desirable dwarf characteristic.
Keywords: DNA variation; auxin; cellulose; dwarf; methylation; soybean; transcriptome.
Copyright © 2022 Song, Wang, Huang, Li, Ren and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis.Theor Appl Genet. 2018 May;131(5):1001-1016. doi: 10.1007/s00122-017-3044-8. Epub 2018 Mar 17. Theor Appl Genet. 2018. PMID: 29550969 Free PMC article.
-
Analysis of genome variants in dwarf soybean lines obtained in F6 derived from cross of normal parents (cultivated and wild soybean).Genomics Inform. 2021 Jun;19(2):e19. doi: 10.5808/gi.21024. Epub 2021 Jun 30. Genomics Inform. 2021. PMID: 34261303 Free PMC article.
-
Transcriptome sequencing reveals hotspot mutation regions and dwarfing mechanisms in wheat mutants induced by γ-ray irradiation and EMS.J Radiat Res. 2020 Jan 23;61(1):44-57. doi: 10.1093/jrr/rrz075. J Radiat Res. 2020. PMID: 31825082 Free PMC article.
-
Analysis of a radiation-induced dwarf mutant of a warm-season turf grass reveals potential mechanisms involved in the dwarfing mutant.Sci Rep. 2020 Nov 3;10(1):18913. doi: 10.1038/s41598-020-75421-x. Sci Rep. 2020. PMID: 33144613 Free PMC article.
-
Comparative transcriptome analysis reveals higher expression of stress and defense responsive genes in dwarf soybeans obtained from the crossing of G. max and G. soja.Genes Genomics. 2019 Nov;41(11):1315-1327. doi: 10.1007/s13258-019-00846-2. Epub 2019 Jul 30. Genes Genomics. 2019. PMID: 31363917
References
-
- Cheng W., Gao J. S., Feng X. X., Shao Q., Yang S. X., Feng X. Z. (2016). Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J. Integr. Agriculture. 15, 2228–2236. doi: 10.1016/S2095-3119(15)61312-0 - DOI
LinkOut - more resources
Full Text Sources

