Nucleo-Cytoplasmic Transport of ZIKV Non-Structural 3 Protein Is Mediated by Importin-α/β and Exportin CRM-1
- PMID: 36475764
- PMCID: PMC9888292
- DOI: 10.1128/jvi.01773-22
Nucleo-Cytoplasmic Transport of ZIKV Non-Structural 3 Protein Is Mediated by Importin-α/β and Exportin CRM-1
Abstract
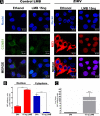
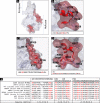
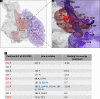
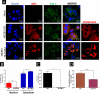
Flaviviruses have a cytoplasmic replicative cycle, and crucial events, such as genome translation and replication, occur in the endoplasmic reticulum. However, some viral proteins, such as C, NS1, and NS5 from Zika virus (ZIKV) containing nuclear localization signals (NLSs) and nuclear export signals (NESs), are also located in the nucleus of Vero cells. The NS2A, NS3, and NS4A proteins from dengue virus (DENV) have also been reported to be in the nucleus of A549 cells, and our group recently reported that the NS3 protein is also located in the nucleus of Huh7 and C636 cells during DENV infection. However, the NS3 protease-helicase from ZIKV locates in the perinuclear region of infected cells and alters the morphology of the nuclear lamina, a component of the nuclear envelope. Furthermore, ZIKV NS3 has been reported to accumulate on the concave face of altered kidney-shaped nuclei and may be responsible for modifying other elements of the nuclear envelope. However, nuclear localization of NS3 from ZIKV has not been substantially investigated in human host cells. Our group has recently reported that DENV and ZIKV NS3 alter the nuclear pore complex (NPC) by cleaving some nucleoporins. Here, we demonstrate the presence of ZIKV NS3 in the nucleus of Huh7 cells early in infection and in the cytoplasm at later times postinfection. In addition, we found that ZIKV NS3 contains an NLS and a putative NES and uses the classic import (importin-α/β) and export pathway via CRM-1 to be transported between the cytoplasm and the nucleus. IMPORTANCE Flaviviruses have a cytoplasmic replication cycle, but recent evidence indicates that nuclear elements play a role in their viral replication. Viral proteins, such as NS5 and C, are imported into the nucleus, and blocking their import prevents replication. Because of the importance of the nucleus in viral replication and the role of NS3 in the modification of nuclear components, we investigated whether NS3 can be localized in the nucleus during ZIKV infection. We found that NS3 is imported into the nucleus via the importin pathway and exported to the cytoplasm via CRM-1. The significance of viral protein nuclear import and export and its relationship with infection establishment is highlighted, emphasizing the development of new host-directed antiviral therapeutic strategies.
Keywords: NES; NLS; ZIKV; flavivirus; nuclear export; nuclear export signal; nuclear localization; nuclear localization signal; viral proteases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Nucleocytoplasmic Shuttling of Porcine Parvovirus NS1 Protein Mediated by the CRM1 Nuclear Export Pathway and the Importin α/β Nuclear Import Pathway.J Virol. 2022 Jan 12;96(1):e0148121. doi: 10.1128/JVI.01481-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643426 Free PMC article.
-
Zika Virus NS5 Forms Supramolecular Nuclear Bodies That Sequester Importin-α and Modulate the Host Immune and Pro-Inflammatory Response in Neuronal Cells.ACS Infect Dis. 2019 Jun 14;5(6):932-948. doi: 10.1021/acsinfecdis.8b00373. Epub 2019 Mar 19. ACS Infect Dis. 2019. PMID: 30848123
-
Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection.Traffic. 2007 Jul;8(7):795-807. doi: 10.1111/j.1600-0854.2007.00579.x. Epub 2007 May 30. Traffic. 2007. PMID: 17537211
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
-
Classical nuclear localization signals: definition, function, and interaction with importin alpha.J Biol Chem. 2007 Feb 23;282(8):5101-5. doi: 10.1074/jbc.R600026200. Epub 2006 Dec 14. J Biol Chem. 2007. PMID: 17170104 Free PMC article. Review.
Cited by
-
An ivermectin - atorvastatin combination impairs nuclear transport inhibiting dengue infection in vitro and in vivo.iScience. 2023 Oct 27;26(12):108294. doi: 10.1016/j.isci.2023.108294. eCollection 2023 Dec 15. iScience. 2023. PMID: 38034354 Free PMC article.
-
The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection.Cells. 2024 Sep 21;13(18):1591. doi: 10.3390/cells13181591. Cells. 2024. PMID: 39329772 Free PMC article. Review.
-
The Disruption of a Nuclear Export Signal in the C-Terminus of the Herpes Simplex Virus 1 Determinant of Pathogenicity UL24 Protein Leads to a Syncytial Plaque Phenotype.Viruses. 2023 Sep 21;15(9):1971. doi: 10.3390/v15091971. Viruses. 2023. PMID: 37766377 Free PMC article.
-
Viral Subversion of the Chromosome Region Maintenance 1 Export Pathway and Its Consequences for the Cell Host.Viruses. 2023 Nov 6;15(11):2218. doi: 10.3390/v15112218. Viruses. 2023. PMID: 38005895 Free PMC article. Review.
-
A CRM1-dependent nuclear export signal in Autographa californica multiple nucleopolyhedrovirus Ac93 is important for the formation of intranuclear microvesicles.J Virol. 2024 May 14;98(5):e0029924. doi: 10.1128/jvi.00299-24. Epub 2024 Apr 1. J Virol. 2024. PMID: 38557225 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

