Evaluation of molecular typing for national surveillance of invasive clinical Haemophilus influenzae isolates from Denmark
- PMID: 36466693
- PMCID: PMC9712784
- DOI: 10.3389/fmicb.2022.1030242
Evaluation of molecular typing for national surveillance of invasive clinical Haemophilus influenzae isolates from Denmark
Abstract
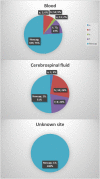
Haemophilus influenzae is a gram-negative coccobacillus known to cause respiratory and invasive infections. It can possess a polysaccharide capsule that can be categorized into six different serotypes (i.e., Hia, Hib, Hic, Hid, Hie, and Hif) and non-encapsulated strains that are defined as non-typeable. Furthermore, H. influenzae can be characterized into eight biotypes (I-VIII). Traditionally, isolates have been serotyped and biotyped using phenotypic methods; however, these methods are not always reliable. In this study, we evaluate the use of whole-genome sequencing (WGS) for national surveillance and characterization of clinical Danish H. influenzae isolates. In Denmark, all clinical invasive isolates between 2014 and 2021 have been serotyped using a traditional phenotypic latex agglutination test as well as in silico serotyped using the in silico programs "hinfluenzae_capsule_characterization" and "hicap" to compare the subsequent serotypes. Moreover, isolates were also biotyped using a phenotypic enzyme test and the genomic data for the detection of the genes encoding ornithine, tryptophan, and urease. The results showed a 99-100% concordance between the two genotypic approaches and the phenotypic serotyping, respectively. The biotyping showed a 95% concordance between genotyping and phenotyping. In conclusion, our results show that in a clinical surveillance setting, in silico serotyping and WGS-based biotyping are a robust and reliable approach for typing clinical H. influenzae isolates.
Keywords: Denmark; Haemophilus influenzae; capsular genes; genotyping; serotyping.
Copyright © 2022 Slotved, Johannesen, Stegger and Fuursted.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Monitoring changes in invasive disease caused by Haemophilus influenzae in the Czech Republic between 1999 and 2020.Epidemiol Mikrobiol Imunol. 2022 Summer;71(2):67-77. Epidemiol Mikrobiol Imunol. 2022. PMID: 35940860 English.
-
Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates.J Clin Microbiol. 2007 Oct;45(10):3230-8. doi: 10.1128/JCM.00794-07. Epub 2007 Aug 15. J Clin Microbiol. 2007. PMID: 17699642 Free PMC article.
-
Trends in invasive Haemophilus influenzae serotype a disease in England from 2008-09 to 2021-22: a prospective national surveillance study.Lancet Infect Dis. 2023 Oct;23(10):1197-1206. doi: 10.1016/S1473-3099(23)00188-3. Epub 2023 Jun 22. Lancet Infect Dis. 2023. PMID: 37356443
-
Haemophilus influenzae serotype a as a cause of serious invasive infections.Lancet Infect Dis. 2014 Jan;14(1):70-82. doi: 10.1016/S1473-3099(13)70170-1. Epub 2013 Nov 20. Lancet Infect Dis. 2014. PMID: 24268829 Review.
-
Invasive Haemophilus influenzae Infections after 3 Decades of Hib Protein Conjugate Vaccine Use.Clin Microbiol Rev. 2021 Jun 16;34(3):e0002821. doi: 10.1128/CMR.00028-21. Epub 2021 Jun 2. Clin Microbiol Rev. 2021. PMID: 34076491 Free PMC article. Review.
Cited by
-
Whole-genome sequencing of non-typeable Haemophilus influenzae isolated from a tertiary care hospital in Surabaya, Indonesia.BMC Infect Dis. 2024 Oct 3;24(1):1097. doi: 10.1186/s12879-024-09826-8. BMC Infect Dis. 2024. PMID: 39358708 Free PMC article.
-
Epidemic Trends and Biofilm Formation Mechanisms of Haemophilus influenzae: Insights into Clinical Implications and Prevention Strategies.Infect Drug Resist. 2023 Aug 16;16:5359-5373. doi: 10.2147/IDR.S424468. eCollection 2023. Infect Drug Resist. 2023. PMID: 37605758 Free PMC article. Review.
-
Development and implementation of a core genome multilocus sequence typing scheme for Haemophilus influenzae.Microb Genom. 2024 Aug;10(8):001281. doi: 10.1099/mgen.0.001281. Microb Genom. 2024. PMID: 39120932 Free PMC article.
-
National Danish surveillance of invasive clinical Haemophilus influenzae isolates and their resistance profile.Front Microbiol. 2023 Nov 22;14:1307261. doi: 10.3389/fmicb.2023.1307261. eCollection 2023. Front Microbiol. 2023. PMID: 38075872 Free PMC article.
References
-
- Fuursted K., Hartmeyer G. N., Stegger M., Andersen P. S., Justesen U. S. (2016). Molecular characterisation of the clonal emergence of high-level ciprofloxacin-monoresistant Haemophilus influenzae in the Region of Southern Denmark. J. Glob. Antimicrob. Resist. 5 67–70. 10.1016/j.jgar.2015.12.004 - DOI - PubMed
LinkOut - more resources
Full Text Sources

