Consequences of Genetic Recombination on Protein Folding Stability
- PMID: 36463317
- PMCID: PMC9849154
- DOI: 10.1007/s00239-022-10080-2
Consequences of Genetic Recombination on Protein Folding Stability
Abstract
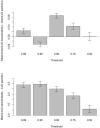
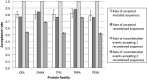
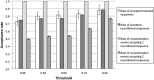
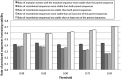
Genetic recombination is a common evolutionary mechanism that produces molecular diversity. However, its consequences on protein folding stability have not attracted the same attention as in the case of point mutations. Here, we studied the effects of homologous recombination on the computationally predicted protein folding stability for several protein families, finding less detrimental effects than we previously expected. Although recombination can affect multiple protein sites, we found that the fraction of recombined proteins that are eliminated by negative selection because of insufficient stability is not significantly larger than the corresponding fraction of proteins produced by mutation events. Indeed, although recombination disrupts epistatic interactions, the mean stability of recombinant proteins is not lower than that of their parents. On the other hand, the difference of stability between recombined proteins is amplified with respect to the parents, promoting phenotypic diversity. As a result, at least one third of recombined proteins present stability between those of their parents, and a substantial fraction have higher or lower stability than those of both parents. As expected, we found that parents with similar sequences tend to produce recombined proteins with stability close to that of the parents. Finally, the simulation of protein evolution along the ancestral recombination graph with empirical substitution models commonly used in phylogenetics, which ignore constraints on protein folding stability, showed that recombination favors the decrease of folding stability, supporting the convenience of adopting structurally constrained models when possible for inferences of protein evolutionary histories with recombination.
Keywords: Molecular evolution; Protein evolution; Protein folding stability; Recombination; Substitution models of protein evolution.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
ProtASR: An Evolutionary Framework for Ancestral Protein Reconstruction with Selection on Folding Stability.Syst Biol. 2017 Nov 1;66(6):1054-1064. doi: 10.1093/sysbio/syw121. Syst Biol. 2017. PMID: 28057858
-
Protein evolution along phylogenetic histories under structurally constrained substitution models.Bioinformatics. 2013 Dec 1;29(23):3020-8. doi: 10.1093/bioinformatics/btt530. Epub 2013 Sep 12. Bioinformatics. 2013. PMID: 24037213 Free PMC article.
-
Maximum-Likelihood Phylogenetic Inference with Selection on Protein Folding Stability.Mol Biol Evol. 2015 Aug;32(8):2195-207. doi: 10.1093/molbev/msv085. Epub 2015 Apr 2. Mol Biol Evol. 2015. PMID: 25837579 Free PMC article.
-
Detecting selection on protein stability through statistical mechanical models of folding and evolution.Biomolecules. 2014 Mar 7;4(1):291-314. doi: 10.3390/biom4010291. Biomolecules. 2014. PMID: 24970217 Free PMC article. Review.
-
Analysing recombination in nucleotide sequences.Mol Ecol Resour. 2011 Nov;11(6):943-55. doi: 10.1111/j.1755-0998.2011.03026.x. Epub 2011 May 19. Mol Ecol Resour. 2011. PMID: 21592314 Review.
Cited by
-
Selection among site-dependent structurally constrained substitution models of protein evolution by approximate Bayesian computation.Bioinformatics. 2024 Mar 4;40(3):btae096. doi: 10.1093/bioinformatics/btae096. Bioinformatics. 2024. PMID: 38374231 Free PMC article.
-
Substitution Models of Protein Evolution with Selection on Enzymatic Activity.Mol Biol Evol. 2024 Feb 1;41(2):msae026. doi: 10.1093/molbev/msae026. Mol Biol Evol. 2024. PMID: 38314876 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

