Cryo-EM structure of ssDNA bacteriophage ΦCjT23 provides insight into early virus evolution
- PMID: 36463224
- PMCID: PMC9719478
- DOI: 10.1038/s41467-022-35123-6
Cryo-EM structure of ssDNA bacteriophage ΦCjT23 provides insight into early virus evolution
Abstract
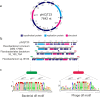
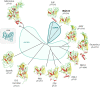
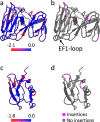
The origin of viruses remains an open question. While lack of detectable sequence similarity hampers the analysis of distantly related viruses, structural biology investigations of conserved capsid protein structures facilitate the study of distant evolutionary relationships. Here we characterize the lipid-containing ssDNA temperate bacteriophage ΦCjT23, which infects Flavobacterium sp. (Bacteroidetes). We report ΦCjT23-like sequences in the genome of strains belonging to several Flavobacterium species. The virion structure determined by cryogenic electron microscopy reveals similarities to members of the viral kingdom Bamfordvirae that currently consists solely of dsDNA viruses with a major capsid protein composed of two upright β-sandwiches. The minimalistic structure of ΦCjT23 suggests that this phage serves as a model for the last common ancestor between ssDNA and dsDNA viruses in the Bamfordvirae. Both ΦCjT23 and the related phage FLiP infect Flavobacterium species found in several environments, suggesting that these types of viruses have a global distribution and a shared evolutionary origin. Detailed comparisons to related, more complex viruses not only expand our knowledge about this group of viruses but also provide a rare glimpse into early virus evolution.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Virus found in a boreal lake links ssDNA and dsDNA viruses.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8378-8383. doi: 10.1073/pnas.1703834114. Epub 2017 Jul 17. Proc Natl Acad Sci U S A. 2017. PMID: 28716906 Free PMC article.
-
Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.mBio. 2022 Aug 30;13(4):e0015622. doi: 10.1128/mbio.00156-22. Epub 2022 Jul 20. mBio. 2022. PMID: 35856561 Free PMC article.
-
ICTV Virus Taxonomy Profile: Finnlakeviridae.J Gen Virol. 2020 Sep;101(9):894-895. doi: 10.1099/jgv.0.001488. J Gen Virol. 2020. PMID: 32840474 Free PMC article.
-
Cryo-EM reveals infection steps of single-stranded RNA bacteriophages.Prog Biophys Mol Biol. 2021 Mar;160:79-86. doi: 10.1016/j.pbiomolbio.2020.07.011. Epub 2020 Aug 22. Prog Biophys Mol Biol. 2021. PMID: 32841651 Review.
-
Major tail proteins of bacteriophages of the order Caudovirales.J Biol Chem. 2022 Jan;298(1):101472. doi: 10.1016/j.jbc.2021.101472. Epub 2021 Dec 8. J Biol Chem. 2022. PMID: 34890646 Free PMC article. Review.
Cited by
-
Jorvik: A membrane-containing phage that will likely found a new family within Vinavirales.iScience. 2023 Sep 29;26(11):108104. doi: 10.1016/j.isci.2023.108104. eCollection 2023 Nov 17. iScience. 2023. PMID: 37867962 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

