Functional proteomic profiling links deficient DNA clearance with increased mortality in individuals with severe COVID-19 pneumonia
- PMID: 36462503
- PMCID: PMC9671605
- DOI: 10.1016/j.immuni.2022.11.007
Functional proteomic profiling links deficient DNA clearance with increased mortality in individuals with severe COVID-19 pneumonia
Abstract
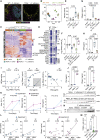
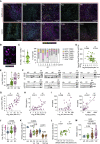
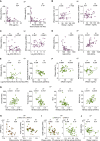
The factors that influence survival during severe infection are unclear. Extracellular chromatin drives pathology, but the mechanisms enabling its accumulation remain elusive. Here, we show that in murine sepsis models, splenocyte death interferes with chromatin clearance through the release of the DNase I inhibitor actin. Actin-mediated inhibition was compensated by upregulation of DNase I or the actin scavenger gelsolin. Splenocyte death and neutrophil extracellular trap (NET) clearance deficiencies were prevalent in individuals with severe COVID-19 pneumonia or microbial sepsis. Activity tracing by plasma proteomic profiling uncovered an association between low NET clearance and increased COVID-19 pathology and mortality. Low NET clearance activity with comparable proteome associations was prevalent in healthy donors with low-grade inflammation, implicating defective chromatin clearance in the development of cardiovascular disease and linking COVID-19 susceptibility to pre-existing conditions. Hence, the combination of aberrant chromatin release with defects in protective clearance mechanisms lead to poor survival outcomes.
Keywords: COVID-19; DNA; DNase I; NETs; actin; degradation; histone; inflammation; proteomics; sepsis.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
Impaired Degradation of Neutrophil Extracellular Traps: A Possible Severity Factor of Elderly Male COVID-19 Patients.J Innate Immun. 2022;14(5):461-476. doi: 10.1159/000521594. Epub 2022 Jan 27. J Innate Immun. 2022. PMID: 35086104 Free PMC article.
-
Hypercholesterolemia Impairs Clearance of Neutrophil Extracellular Traps and Promotes Inflammation and Atherosclerotic Plaque Progression.Arterioscler Thromb Vasc Biol. 2021 Oct;41(10):2598-2615. doi: 10.1161/ATVBAHA.120.316389. Epub 2021 Aug 5. Arterioscler Thromb Vasc Biol. 2021. PMID: 34348488 Free PMC article.
-
Neutrophil extracellular trap inhibition improves survival in neonatal mouse infectious peritonitis.Pediatr Res. 2023 Mar;93(4):862-869. doi: 10.1038/s41390-022-02219-0. Epub 2022 Jul 28. Pediatr Res. 2023. PMID: 35902703 Free PMC article.
-
Therapeutic Targeting of Neutrophil Extracellular Traps in Atherogenic Inflammation.Thromb Haemost. 2019 Apr;119(4):542-552. doi: 10.1055/s-0039-1678664. Epub 2019 Feb 7. Thromb Haemost. 2019. PMID: 30731493 Review.
-
Periodontitis-Derived Dark-NETs in Severe Covid-19.Front Immunol. 2022 Apr 12;13:872695. doi: 10.3389/fimmu.2022.872695. eCollection 2022. Front Immunol. 2022. PMID: 35493525 Free PMC article. Review.
Cited by
-
Development of a Clonal and High-Yield Mammalian Cell Line for the Manufacturing of a Hyperactive Human DNase I with Extended Plasma Half-Life Using PASylation® Technology.Pharmaceutics. 2024 Jul 22;16(7):967. doi: 10.3390/pharmaceutics16070967. Pharmaceutics. 2024. PMID: 39065664 Free PMC article.
-
Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation.Int J Mol Sci. 2023 Feb 10;24(4):3570. doi: 10.3390/ijms24043570. Int J Mol Sci. 2023. PMID: 36834989 Free PMC article.
-
Impaired balance between neutrophil extracellular trap formation and degradation by DNases in COVID-19 disease.J Transl Med. 2024 Mar 7;22(1):246. doi: 10.1186/s12967-024-05044-7. J Transl Med. 2024. PMID: 38454482 Free PMC article.
-
You are what you eat and how you digest it! A discussion on inflammatory efferocytosis.Front Cell Dev Biol. 2023 Feb 9;11:1132696. doi: 10.3389/fcell.2023.1132696. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36846584 Free PMC article. Review.
-
Visualization of Nuclease- and Serum-Mediated Chromatin Degradation with DNA-Histone Mesostructures.Int J Mol Sci. 2023 Feb 6;24(4):3222. doi: 10.3390/ijms24043222. Int J Mol Sci. 2023. PMID: 36834634 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- FC0010129/CRUK_/Cancer Research UK/United Kingdom
- FC001134 /CRUK_/Cancer Research UK/United Kingdom
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- PG/18/45/33814/BHF_/British Heart Foundation/United Kingdom
- FC001134 /WT_/Wellcome Trust/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- FC0010129/MRC_/Medical Research Council/United Kingdom
- FC001134/ARC_/Arthritis Research UK/United Kingdom
- MC_PC_19009/MRC_/Medical Research Council/United Kingdom
- 222825/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- FC0010129/WT_/Wellcome Trust/United Kingdom
- FC001134 /MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials

