Diagnostic implication of a circulating serum-based three-microRNA signature in hepatocellular carcinoma
- PMID: 36457743
- PMCID: PMC9705795
- DOI: 10.3389/fgene.2022.929787
Diagnostic implication of a circulating serum-based three-microRNA signature in hepatocellular carcinoma
Abstract
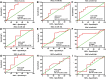
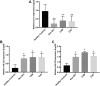
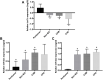
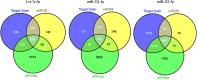
Owing to the diagnostic dilemma, the prognosis of hepatocellular carcinoma (HCC) remains impoverished, contributing to the globally high mortality rate. Currently, HCC diagnosis depends on the combination of imaging modalities and the measurement of serum alpha-fetoprotein (AFP) levels. Nevertheless, these conventional modalities exhibit poor performance in detecting HCC at early stages. Thus, there is a pressing need to identify novel circulating biomarkers to promote diagnostic accuracy and surveillance. Circulating miRNAs are emerging as promising diagnostic tools in screening various cancers, including HCC. However, because of heterogenous and, at times, contradictory reports, the universality of miRNAs in clinical settings remains elusive. Consequently, we proposed to explore the diagnostic potential of ten miRNAs selected on a candidate-based approach in HCC diagnosis. The expression of ten candidate miRNAs (Let-7a, miR-15a, miR-26a, miR-124, miR-126, miR-155, miR-219, miR-221, miR-222, and miR-340) was investigated in serum and tissue of 66 subjects, including 33 HCC patients and 33 healthy controls (HC), by rt-PCR. Receiver operating characteristic curve (ROC) analysis was used to determine the diagnostic accuracy of the prospective serum miRNA panel. To anticipate the potential biological roles of a three-miRNA signature, the target genes were evaluated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway. The serum and tissue expression of miRNAs (Let-7a, miR-26a, miR-124, miR-155, miR-221, miR-222, and miR-340) were differentially expressed in HCC patients (p < 0.05). The ROC analysis revealed promising diagnostic performance of Let-7a (AUC = 0.801), miR-221 (AUC = 0.786), and miR-2 (AUC = 0.758) in discriminating HCC from HC. Furthermore, in a logistic regression equation, we identified a three-miRNA panel (Let-7a, miR-221, and miR-222; AUC = 0.932) with improved diagnostic efficiency in differentiating HCC from HC. Remarkably, the combination of AFP and a three-miRNA panel offered a higher accuracy of HCC diagnosis (AUC = 0.961) than AFP alone. The functional enrichment analysis demonstrated that target genes may contribute to pathways associated with HCC and cell-cycle regulation, indicating possible crosstalk of miRNAs with HCC development. To conclude, the combined classifier of a three-miRNA panel and AFP could be indispensable circulating biomarkers for HCC diagnosis. Furthermore, targeting predicted genes may provide new therapeutic clues for the treatment of aggressive HCC.
Keywords: biomarker; circulating miRNA; diagnosis; hepatocellular carcinoma; therapeutics.
Copyright © 2022 Yousuf, Dar, Bangri, Choh, Rasool, Shah, Rather, Rah, Bhat, Ali and Afroze.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Optimisation of quantitative miRNA panels to consolidate the diagnostic surveillance of HBV-related hepatocellular carcinoma.PLoS One. 2018 Apr 19;13(4):e0196081. doi: 10.1371/journal.pone.0196081. eCollection 2018. PLoS One. 2018. PMID: 29672637 Free PMC article.
-
miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma.Oncol Lett. 2019 Sep;18(3):3381-3387. doi: 10.3892/ol.2019.10606. Epub 2019 Jul 11. Oncol Lett. 2019. PMID: 31452818 Free PMC article.
-
Development and Validation of a Novel Circulating miRNA-Based Diagnostic Score for Early Detection of Hepatocellular Carcinoma.Dig Dis Sci. 2022 Jun;67(6):2283-2292. doi: 10.1007/s10620-021-07031-0. Epub 2021 May 12. Dig Dis Sci. 2022. PMID: 33982217
-
Circulating microRNAs for the diagnosis of hepatocellular carcinoma.Dig Liver Dis. 2019 May;51(5):621-631. doi: 10.1016/j.dld.2018.12.011. Epub 2018 Dec 28. Dig Liver Dis. 2019. PMID: 30744930 Review.
-
Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis.Clin Transl Oncol. 2015 Sep;17(9):684-93. doi: 10.1007/s12094-015-1294-y. Epub 2015 May 9. Clin Transl Oncol. 2015. PMID: 25956842 Review.
Cited by
-
What are the changes in the hotspots and frontiers of microRNAs in hepatocellular carcinoma over the past decade?World J Clin Oncol. 2024 Jan 24;15(1):145-158. doi: 10.5306/wjco.v15.i1.145. World J Clin Oncol. 2024. PMID: 38292666 Free PMC article.
-
A liquid biopsy assay for the noninvasive detection of lymph node metastases in T1 lung adenocarcinoma.Thorac Cancer. 2024 Jun;15(16):1312-1319. doi: 10.1111/1759-7714.15315. Epub 2024 Apr 29. Thorac Cancer. 2024. PMID: 38682829 Free PMC article.
-
Circulating microRNAs as promising diagnostic biomarkers for hepatocellular carcinoma: a systematic review and meta-analysis.Front Mol Biosci. 2024 May 14;11:1353547. doi: 10.3389/fmolb.2024.1353547. eCollection 2024. Front Mol Biosci. 2024. PMID: 38808007 Free PMC article. Review.
-
N7-methylguanosine-related miRNAs predict hepatocellular carcinoma prognosis and immune therapy.Aging (Albany NY). 2023 Nov 3;15(21):12192-12208. doi: 10.18632/aging.205172. Epub 2023 Nov 3. Aging (Albany NY). 2023. PMID: 37925170 Free PMC article.
-
Serum microRNA Profiles and Pathways in Hepatitis B-Associated Hepatocellular Carcinoma: A South African Study.Int J Mol Sci. 2024 Jan 12;25(2):975. doi: 10.3390/ijms25020975. Int J Mol Sci. 2024. PMID: 38256049 Free PMC article. Review.
References
-
- Acunzo M., Romano G., Wernicke D., Croce C. M. (2015). MicroRNA and cancer--a brief overview. Adv. Biol. Regul. 57, 1–9. - PubMed
-
- Aleman S., Rahbin N., Weiland O., Davidsdottir L., Hedenstierna M., Rose N., et al. (2013). A risk for hepatocellular carcinoma persists long-term after sustained virologic response in patients with hepatitis C-associated liver cirrhosis. Clin. Infect. Dis. 57, 230–236. - PubMed
LinkOut - more resources
Full Text Sources

