Proteomic Comparison of Three Wild-Type Pseudorabies Virus Strains and the Attenuated Bartha Strain Reveals Reduced Incorporation of Several Tegument Proteins in Bartha Virions
- PMID: 36453884
- PMCID: PMC9769387
- DOI: 10.1128/jvi.01158-22
Proteomic Comparison of Three Wild-Type Pseudorabies Virus Strains and the Attenuated Bartha Strain Reveals Reduced Incorporation of Several Tegument Proteins in Bartha Virions
Abstract
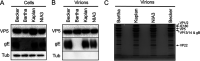
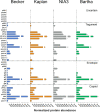
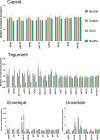
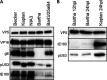
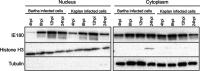
Pseudorabies virus (PRV) is a member of the alphaherpesvirus subfamily and the causative agent of Aujeszky's disease in pigs. Driven by the large economic losses associated with PRV infection, several vaccines and vaccine programs have been developed. To this day, the attenuated Bartha strain, generated by serial passaging, represents the golden standard for PRV vaccination. However, a proteomic comparison of the Bartha virion to wild-type (WT) PRV virions is lacking. Here, we present a comprehensive mass spectrometry-based proteome comparison of the attenuated Bartha strain and three commonly used WT PRV strains: Becker, Kaplan, and NIA3. We report the detection of 40 structural and 14 presumed nonstructural proteins through a combination of data-dependent and data-independent acquisition. Interstrain comparisons revealed that packaging of the capsid and most envelope proteins is largely comparable in-between all four strains, except for the envelope protein pUL56, which is less abundant in Bartha virions. However, distinct differences were noted for several tegument proteins. Most strikingly, we noted a severely reduced incorporation of the tegument proteins IE180, VP11/12, pUS3, VP22, pUL41, pUS1, and pUL40 in Bartha virions. Moreover, and likely as a consequence, we also observed that Bartha virions are on average smaller and more icosahedral compared to WT virions. Finally, we detected at least 28 host proteins that were previously described in PRV virions and noticed considerable strain-specific differences with regard to host proteins, arguing that the potential role of packaged host proteins in PRV replication and spread should be further explored. IMPORTANCE The pseudorabies virus (PRV) vaccine strain Bartha-an attenuated strain created by serial passaging-represents an exceptional success story in alphaherpesvirus vaccination. Here, we used mass spectrometry to analyze the Bartha virion composition in comparison to three established WT PRV strains. Many viral tegument proteins that are considered nonessential for viral morphogenesis were drastically less abundant in Bartha virions compared to WT virions. Interestingly, many of the proteins that are less incorporated in Bartha participate in immune evasion strategies of alphaherpesviruses. In addition, we observed a reduced size and more icosahedral morphology of the Bartha virions compared to WT PRV. Given that the Bartha vaccine strain elicits potent immune responses, our findings here suggest that differences in protein packaging may contribute to its immunogenicity. Further exploration of these observations could aid the development of efficacious vaccines against other alphaherpesvirus vaccines such as HSV-1/2 or EHV-1.
Keywords: Aujeszky’s disease virus; Bartha; Becker; Kaplan; NIA3; PRV; data-dependent acquisition; data-independent acquisition; mass spectrometry; proteome; proteomics; pseudorabies virus; suid herpesvirus 1; tegument.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Attenuated Pseudorabies Virus Vaccine Strain Bartha Hyperactivates Plasmacytoid Dendritic Cells by Generating Large Amounts of Cell-Free Virus in Infected Epithelial Cells.J Virol. 2022 Jun 22;96(12):e0219921. doi: 10.1128/jvi.02199-21. Epub 2022 May 23. J Virol. 2022. PMID: 35604216 Free PMC article.
-
Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21.J Virol. 2007 Jan;81(2):1048-51. doi: 10.1128/JVI.01801-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079290 Free PMC article.
-
Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge.Vaccine. 2015 Oct 26;33(43):5733-5740. doi: 10.1016/j.vaccine.2015.09.066. Epub 2015 Oct 1. Vaccine. 2015. PMID: 26428456
-
The Attenuated Pseudorabies Virus Vaccine Strain Bartha K61: A Brief Review on the Knowledge Gathered During 60 Years of Research.Pathogens. 2020 Oct 27;9(11):897. doi: 10.3390/pathogens9110897. Pathogens. 2020. PMID: 33121171 Free PMC article. Review.
-
Vaccines against pseudorabies virus (PrV).Vet Microbiol. 2017 Jul;206:3-9. doi: 10.1016/j.vetmic.2016.11.019. Epub 2016 Nov 18. Vet Microbiol. 2017. PMID: 27890448 Review.
Cited by
-
Virus-Host Protein Interaction Network of the Hepatitis E Virus ORF2-4 by Mammalian Two-Hybrid Assays.Viruses. 2023 Dec 12;15(12):2412. doi: 10.3390/v15122412. Viruses. 2023. PMID: 38140653 Free PMC article.
References
-
- Pellett PM, Roizman B. 2001. The family Herpesviridae: a brief introduction. In Fields virology, 5th ed. Wolters Kluwer Health, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

