Subtle Longitudinal Alterations in Env Sequence Potentiate Differences in Sensitivity to Broadly Neutralizing Antibodies following Acute HIV-1 Subtype C Infection
- PMID: 36453881
- PMCID: PMC9769376
- DOI: 10.1128/jvi.01270-22
Subtle Longitudinal Alterations in Env Sequence Potentiate Differences in Sensitivity to Broadly Neutralizing Antibodies following Acute HIV-1 Subtype C Infection
Abstract
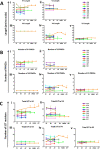
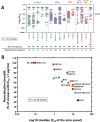
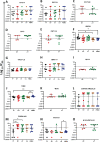
Broadly neutralizing antibodies (bNAbs) for HIV-1 prevention or cure strategies must inhibit transmitted/founder and reservoir viruses. Establishing sensitivity of circulating viruses to bNAbs and genetic patterns affecting neutralization variability may guide rational bNAbs selection for clinical development. We analyzed 326 single env genomes from nine individuals followed longitudinally following acute HIV-1 infection, with samples collected at ~1 week after the first detection of plasma viremia; 300 to 1,709 days postinfection but prior to initiating antiretroviral therapy (ART) (median = 724 days); and ~1 year post ART initiation. Sequences were assessed for phylogenetic relatedness, potential N- and O-linked glycosylation, and variable loop lengths (V1 to V5). A total of 43 env amplicons (median = 3 per patient per time point) were cloned into an expression vector and the TZM-bl assay was used to assess the neutralization profiles of 15 bNAbs targeting the CD4 binding site, V1/V2 region, V3 supersite, MPER, gp120/gp41 interface, and fusion peptide. At 1 μg/mL, the neutralization breadths were as follows: VRC07-LS and N6.LS (100%), VRC01 (86%), PGT151 (81%), 10-1074 and PGT121 (80%), and less than 70% for 10E8, 3BNC117, CAP256.VRC26, 4E10, PGDM1400, and N123-VRC34.01. Features associated with low sensitivity to V1/V2 and V3 bNAbs were higher potential glycosylation sites and/or relatively longer V1 and V4 domains, including known "signature" mutations. The study shows significant variability in the breadth and potency of bNAbs against circulating HIV-1 subtype C envelopes. VRC07-LS, N6.LS, VRC01, PGT151, 10-1074, and PGT121 display broad activity against subtype C variants, and major determinants of sensitivity to most bNAbs were within the V1/V4 domains. IMPORTANCE Broadly neutralizing antibodies (bNAbs) have potential clinical utility in HIV-1 prevention and cure strategies. However, bNAbs target diverse epitopes on the HIV-1 envelope and the virus may evolve to evade immune responses. It is therefore important to identify antibodies with broad activity in high prevalence settings, as well as the genetic patterns that may lead to neutralization escape. We investigated 15 bNAbs with diverse biophysical properties that target six epitopes of the HIV-1 Env glycoprotein for their ability to inhibit viruses that initiated infection, viruses circulating in plasma at chronic infection before antiretroviral treatment (ART), or viruses that were archived in the reservoir during ART in subtype C infected individuals in South Africa, a high burden country. We identify the antibodies most likely to be effective for clinical use in this setting and describe mutational patterns associated with neutralization escape from these antibodies.
Keywords: HIV-1 envelope; HIV-1 intraparticipant diversity; HIV-1 subtype C; acute HIV-1 infection; bNAbs; longitudinal HIV-1 diversity; sensitivity to bNAbs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Sensitivity to Broadly Neutralizing Antibodies of Recently Transmitted HIV-1 Clade CRF02_AG Viruses with a Focus on Evolution over Time.J Virol. 2019 Jan 4;93(2):e01492-18. doi: 10.1128/JVI.01492-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404804 Free PMC article.
-
Long-Term and Low-Level Envelope C2V3 Stimulation by Highly Diverse Virus Isolates Leads to Frequent Development of Broad and Elite Antibody Neutralization in HIV-1-Infected Individuals.Microbiol Spectr. 2022 Dec 21;10(6):e0163422. doi: 10.1128/spectrum.01634-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445130 Free PMC article.
-
Characterizing the Relationship Between Neutralization Sensitivity and env Gene Diversity During ART Suppression.Front Immunol. 2021 Sep 15;12:710327. doi: 10.3389/fimmu.2021.710327. eCollection 2021. Front Immunol. 2021. PMID: 34603284 Free PMC article.
-
Broadly neutralizing antibodies for the treatment and prevention of HIV infection.Curr Opin HIV AIDS. 2020 Jan;15(1):49-55. doi: 10.1097/COH.0000000000000600. Curr Opin HIV AIDS. 2020. PMID: 31764199 Free PMC article. Review.
-
Advancing HIV Broadly Neutralizing Antibodies: From Discovery to the Clinic.Front Public Health. 2021 May 26;9:690017. doi: 10.3389/fpubh.2021.690017. eCollection 2021. Front Public Health. 2021. PMID: 34123998 Free PMC article. Review.
Cited by
-
Predicted resistance to broadly neutralizing antibodies (bnAbs) and associated HIV-1 envelope characteristics among seroconverting adults in Botswana.Sci Rep. 2023 Oct 24;13(1):18134. doi: 10.1038/s41598-023-44722-2. Sci Rep. 2023. PMID: 37875518 Free PMC article.
-
HIV-1 envelope facilitates the development of protease inhibitor resistance through acquiring mutations associated with viral entry and immune escape.Front Microbiol. 2024 Apr 18;15:1388729. doi: 10.3389/fmicb.2024.1388729. eCollection 2024. Front Microbiol. 2024. PMID: 38699474 Free PMC article.
References
-
- McFarland EJ, Cunningham CK, Muresan P, Capparelli EV, Perlowski C, Morgan P, Smith B, Hazra R, Purdue L, Harding PA, Theron G, Mujuru H, Agwu A, Purswani M, Rathore MH, Flach B, Taylor A, Lin BC, McDermott AB, Mascola JR, Graham BS, Rossouw M, Rossouw L, Louw J, Vhembo T, Mhembere TP, Matibe P, Mahmoudi S, Maldonado A, Maraqa N, Baig MM, Rogo T, Cavallo M, Collinson-Streng A, Anderson T, Golden WC, Persaud D, Puga AM, Robinson L-G, Eysallenne Z, Leon D, Paul ME, McMullen-Jackson C, Buschur S, Pontifes M, Sung J, Glenny C, Dunn J, Navarro K, International Maternal Pediatric Adolescent AIDS Clinical Trials Network (IMPAACT) P1112 Team . 2021. Safety, tolerability, and pharmacokinetics of a long-acting broadly neutralizing HIV-1 monoclonal antibody VRC01LS in HIV-1-exposed newborn infants. The J Infectious Diseases 224:1916–1924. 10.1093/infdis/jiab229. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

