Systematic generation and imaging of tandem repeats reveal base-pairing properties that promote RNA aggregation
- PMID: 36452875
- PMCID: PMC9701603
- DOI: 10.1016/j.crmeth.2022.100334
Systematic generation and imaging of tandem repeats reveal base-pairing properties that promote RNA aggregation
Abstract
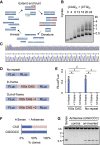
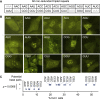
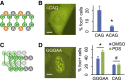
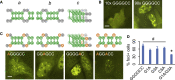
A common pathological feature of RNAs containing expanded repeat sequences is their propensity to aggregate in cells. While some repeat RNA aggregates have been shown to cause toxicity by sequestering RNA-binding proteins, the molecular mechanism of aggregation remains unclear. Here, we devised an efficient method to generate long tandem repeat DNAs de novo and applied it to systematically determine the sequence features underlying RNA aggregation. Live-cell imaging of repeat RNAs indicated that aggregation was promoted by multivalent RNA-RNA interactions via either canonical or noncanonical base pairs. While multiple runs of two consecutive base pairs were sufficient, longer runs of base pairs such as those formed by GGGGCC hexanucleotide repeats further enhanced aggregation. In summary, our study provides a unifying model for the molecular basis of repeat RNA aggregation and a generalizable approach for identifying the sequence and structural determinants underlying the distinct properties of repeat DNAs and RNAs.
Keywords: G-quadruplex; RNA aggregation; RNA base-pairing; RNA imaging; RNA structure; microsatellite; multivalent interactions; repeat expansion disorder; short tandem repeat.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures.J Biol Chem. 2013 Apr 5;288(14):9860-9866. doi: 10.1074/jbc.C113.452532. Epub 2013 Feb 19. J Biol Chem. 2013. PMID: 23423380 Free PMC article.
-
RNA phase transitions in repeat expansion disorders.Nature. 2017 Jun 8;546(7657):243-247. doi: 10.1038/nature22386. Epub 2017 May 31. Nature. 2017. PMID: 28562589 Free PMC article.
-
TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins.J Biol Chem. 2014 Feb 21;289(8):4653-9. doi: 10.1074/jbc.C113.502336. Epub 2013 Dec 26. J Biol Chem. 2014. PMID: 24371143 Free PMC article.
-
Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners.Molecules. 2023 Aug 1;28(15):5801. doi: 10.3390/molecules28155801. Molecules. 2023. PMID: 37570771 Free PMC article. Review.
-
Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration.Neurobiol Aging. 2014 Oct;35(10):2421.e1-2421.e12. doi: 10.1016/j.neurobiolaging.2014.04.015. Epub 2014 Apr 19. Neurobiol Aging. 2014. PMID: 24836899 Review.
Cited by
-
Structural investigation of pathogenic RFC1 AAGGG pentanucleotide repeats reveals a role of G-quadruplex in dysregulated gene expression in CANVAS.Nucleic Acids Res. 2024 Mar 21;52(5):2698-2710. doi: 10.1093/nar/gkae032. Nucleic Acids Res. 2024. PMID: 38266156 Free PMC article.
-
Odd-even disparity in the population of slipped hairpins in RNA repeat sequences with implications for phase separation.Proc Natl Acad Sci U S A. 2023 Jun 13;120(24):e2301409120. doi: 10.1073/pnas.2301409120. Epub 2023 Jun 5. Proc Natl Acad Sci U S A. 2023. PMID: 37276412 Free PMC article.
-
Aberrant splicing exonizes C9ORF72 repeat expansion in ALS/FTD.bioRxiv [Preprint]. 2023 Nov 14:2023.11.13.566896. doi: 10.1101/2023.11.13.566896. bioRxiv. 2023. PMID: 38014069 Free PMC article. Preprint.
References
-
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

