m6A-modified circFNDC3B inhibits colorectal cancer stemness and metastasis via RNF41-dependent ASB6 degradation
- PMID: 36446779
- PMCID: PMC9709059
- DOI: 10.1038/s41419-022-05451-y
m6A-modified circFNDC3B inhibits colorectal cancer stemness and metastasis via RNF41-dependent ASB6 degradation
Abstract
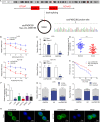
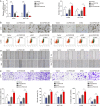
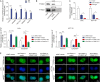
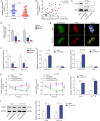
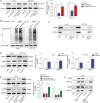
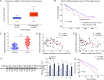
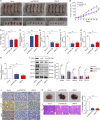
Colorectal cancer (CRC) is the third most frequently diagnosed cancer with unfavorable clinical outcomes worldwide. circFNDC3B plays as a tumor suppressor in CRC, however, the mechanism of circFNDC3B in CRC remains ambiguous. The stem-like properties of CRC cells were detected by the evaluation of stemness markers, sphere formation assay and flow cytometry. qRT-PCR, FISH, IHC, and western blotting assessed the expression and localization of circFNDC3B, RNF41, ASB6, and stemness markers in CRC. The metastatic capabilities of CRC cells were examined by wound healing and Transwell assays, as well as in vivo liver metastasis model. Bioinformatics analysis, RNA immunoprecipitation (RIP), RNA pull-down assay and co-IP were used to detect the associations among circFNDC3B, FXR2, RNF41, and ASB6. Downregulated circFNDC3B was associated with unfavorite survival in CRC patients, and circFNDC3B overexpression suppressed CRC stemness and metastasis. Mechanistically, studies revealed that YTHDC1 facilitated cytoplasmic translocation of m6A-modified circFNDC3B, and circFNDC3B enhanced RNF41 mRNA stability and expression via binding to FXR2. circFNDC3B promoted ASB6 degradation through RNF41-mediated ubiquitination. Functional studies showed that silencing of RNF41 counteracted circFNDC3B-suppressed CRC stemness and metastasis, and ASB6 overexpression reversed circFNDC3B- or RNF41-mediated regulation of CRC stemness and metastasis. Elevated ASB6 was positively correlated with unfavorite survival in CRC patients. In vivo experiments further showed that circFNDC3B or RNF41 overexpression repressed tumor growth, stemness and liver metastasis via modulating ASB6. Taken together, m6A-modified circFNDC3B inhibited CRC stemness and metastasis via RNF41-dependent ASB6 degradation. These findings provide novel insights and important clues for targeted therapeutic strategies of CRC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression.Mol Oncol. 2020 Nov;14(11):2960-2984. doi: 10.1002/1878-0261.12796. Epub 2020 Sep 19. Mol Oncol. 2020. PMID: 32896063 Free PMC article.
-
m6A modified BACE1-AS contributes to liver metastasis and stemness-like properties in colorectal cancer through TUFT1 dependent activation of Wnt signaling.J Exp Clin Cancer Res. 2023 Nov 21;42(1):306. doi: 10.1186/s13046-023-02881-0. J Exp Clin Cancer Res. 2023. PMID: 37986103 Free PMC article.
-
Ubiquitin-specific peptidase 15 regulates the TFAP4/PCGF1 axis facilitating liver metastasis of colorectal cancer and cell stemness.Biochem Pharmacol. 2024 Aug;226:116319. doi: 10.1016/j.bcp.2024.116319. Epub 2024 May 25. Biochem Pharmacol. 2024. PMID: 38801926
-
Emerging mechanisms progress of colorectal cancer liver metastasis.Front Endocrinol (Lausanne). 2022 Dec 8;13:1081585. doi: 10.3389/fendo.2022.1081585. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36568117 Free PMC article. Review.
-
Emerging landscape of circFNDC3B and its role in human malignancies.Front Oncol. 2023 Jan 31;13:1097956. doi: 10.3389/fonc.2023.1097956. eCollection 2023. Front Oncol. 2023. PMID: 36793611 Free PMC article. Review.
Cited by
-
PCMT1 regulates the migration, invasion, and apoptosis of prostate cancer through modulating the PI3K/AKT/GSK-3β pathway.Aging (Albany NY). 2023 Oct 27;15(20):11654-11671. doi: 10.18632/aging.205152. Epub 2023 Oct 27. Aging (Albany NY). 2023. PMID: 37899170 Free PMC article.
-
m6A modification on the fate of colorectal cancer: functions and mechanisms of cell proliferation and tumorigenesis.Front Oncol. 2023 Apr 21;13:1162300. doi: 10.3389/fonc.2023.1162300. eCollection 2023. Front Oncol. 2023. PMID: 37152066 Free PMC article. Review.
-
Interaction of the intestinal cytokines-JAKs-STAT3 and 5 axes with RNA N6-methyladenosine to promote chronic inflammation-induced colorectal cancer.Front Oncol. 2024 Jul 29;14:1352845. doi: 10.3389/fonc.2024.1352845. eCollection 2024. Front Oncol. 2024. PMID: 39136000 Free PMC article. Review.
-
Circular RNAs-New Kids on the Block in Cancer Pathophysiology and Management.Cells. 2023 Feb 8;12(4):552. doi: 10.3390/cells12040552. Cells. 2023. PMID: 36831219 Free PMC article. Review.
-
circFNDC3B Accelerates Vasculature Formation and Metastasis in Oral Squamous Cell Carcinoma.Cancer Res. 2023 May 2;83(9):1459-1475. doi: 10.1158/0008-5472.CAN-22-2585. Cancer Res. 2023. PMID: 36811957 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

