Next-generation retinoid X receptor agonists increase ATRA signaling in organotypic epithelium cultures and have distinct effects on receptor dynamics
- PMID: 36436565
- PMCID: PMC9807999
- DOI: 10.1016/j.jbc.2022.102746
Next-generation retinoid X receptor agonists increase ATRA signaling in organotypic epithelium cultures and have distinct effects on receptor dynamics
Abstract
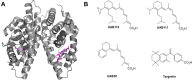
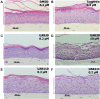
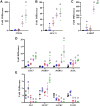
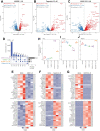
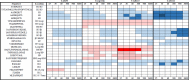
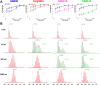
Retinoid X receptors (RXRs) are nuclear transcription factors that partner with other nuclear receptors to regulate numerous physiological processes. Although RXR represents a valid therapeutic target, only a few RXR-specific ligands (rexinoids) have been identified, in part due to the lack of clarity on how rexinoids selectively modulate RXR response. Previously, we showed that rexinoid UAB30 potentiates all-trans-retinoic acid (ATRA) signaling in human keratinocytes, in part by stimulating ATRA biosynthesis. Here, we examined the mechanism of action of next-generation rexinoids UAB110 and UAB111 that are more potent in vitro than UAB30 and the FDA-approved Targretin. Both UAB110 and UAB111 enhanced ATRA signaling in human organotypic epithelium at a 50-fold lower concentration than UAB30. This was consistent with the 2- to 5- fold greater increase in ATRA in organotypic epidermis treated with UAB110/111 versus UAB30. Furthermore, at 0.2 μM, UAB110/111 increased the expression of ATRA genes up to 16-fold stronger than Targretin. The less toxic and more potent UAB110 also induced more changes in differential gene expression than Targretin. Additionally, our hydrogen deuterium exchange mass spectrometry analysis showed that both ligands reduced the dynamics of the ligand-binding pocket but also induced unique dynamic responses that were indicative of higher affinity binding relative to UAB30, especially for Helix 3. UAB110 binding also showed increased dynamics towards the dimer interface through the Helix 8 and Helix 9 regions. These data suggest that UAB110 and UAB111 are potent activators of RXR-RAR signaling pathways but accomplish activation through different molecular responses to ligand binding.
Keywords: Targretin; UAB110; UAB111; UAB30; nuclear receptor; organotypic epidermis; retinoic acid; retinoid X receptor; rexinoid agonists.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
The retinoid X receptor has a critical role in synthetic rexinoid-induced increase in cellular all-trans-retinoic acid.PLoS One. 2024 Apr 1;19(4):e0301447. doi: 10.1371/journal.pone.0301447. eCollection 2024. PLoS One. 2024. PMID: 38557762 Free PMC article.
-
Defining the communication between agonist and coactivator binding in the retinoid X receptor α ligand binding domain.J Biol Chem. 2014 Jan 10;289(2):814-26. doi: 10.1074/jbc.M113.476861. Epub 2013 Nov 1. J Biol Chem. 2014. PMID: 24187139 Free PMC article.
-
Retinoid X Receptor Agonists Upregulate Genes Responsible for the Biosynthesis of All-Trans-Retinoic Acid in Human Epidermis.PLoS One. 2016 Apr 14;11(4):e0153556. doi: 10.1371/journal.pone.0153556. eCollection 2016. PLoS One. 2016. PMID: 27078158 Free PMC article.
-
Modulation of RXR function through ligand design.Biochim Biophys Acta. 2012 Jan;1821(1):57-69. doi: 10.1016/j.bbalip.2011.04.003. Epub 2011 Apr 16. Biochim Biophys Acta. 2012. PMID: 21515403 Review.
-
Natural and Structure-based RXR Ligand Scaffolds and Their Functions.Curr Top Med Chem. 2017;17(6):631-662. doi: 10.2174/1568026616666160617072521. Curr Top Med Chem. 2017. PMID: 27320335 Review.
Cited by
-
The retinoid X receptor has a critical role in synthetic rexinoid-induced increase in cellular all-trans-retinoic acid.PLoS One. 2024 Apr 1;19(4):e0301447. doi: 10.1371/journal.pone.0301447. eCollection 2024. PLoS One. 2024. PMID: 38557762 Free PMC article.
-
Modeling Epithelial Homeostasis and Perturbation in Three-Dimensional Human Esophageal Organoids.Biomolecules. 2024 Sep 5;14(9):1126. doi: 10.3390/biom14091126. Biomolecules. 2024. PMID: 39334892 Free PMC article.
-
Epidermal retinol dehydrogenases cyclically regulate stem cell markers and clock genes and influence hair composition.Commun Biol. 2024 Apr 12;7(1):453. doi: 10.1038/s42003-024-06160-2. Commun Biol. 2024. PMID: 38609439 Free PMC article.
-
Retinoid X Receptor agonists as selective modulators of the immune system for the treatment of cancer.Pharmacol Ther. 2023 Dec;252:108561. doi: 10.1016/j.pharmthera.2023.108561. Epub 2023 Nov 10. Pharmacol Ther. 2023. PMID: 37952906 Free PMC article. Review.
References
-
- Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. - PubMed
-
- Ottow E., Weinmann H. Nuclear receptors as drug targets: a historical perspective of modern drug discovery. Nucl. Receptors as Drug Targets. 2008;7:679–684.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

