Single-cell multiomics reveals the complexity of TGFβ signalling to chromatin in iPSC-derived kidney organoids
- PMID: 36435939
- PMCID: PMC9701233
- DOI: 10.1038/s42003-022-04264-1
Single-cell multiomics reveals the complexity of TGFβ signalling to chromatin in iPSC-derived kidney organoids
Abstract
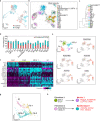
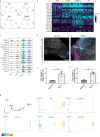
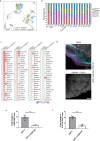
TGFβ1 plays a regulatory role in the determination of renal cell fate and the progression of renal fibrosis. Here we show an association between SMAD3 and the histone methyltransferase, EZH2, during cell differentiation; ChIP-seq revealed that SMAD3 and EZH2 co-occupy the genome in iPSCs and in iPSC-derived nephron progenitors. Through integration of single cell gene expression and epigenome profiling, we identified de novo ACTA2+ve/POSTN+ve myofibroblasts in kidney organoids treated with TGFβ1, characterised by increased SMAD3-dependent cis chromatin accessibility and gene expression associated with fibroblast activation. We have identified fibrosis-associated regulons characterised by enrichment of SMAD3, AP1, the ETS family of transcription factors, and NUAK1, CREB3L1, and RARG, corresponding to enriched motifs at accessible loci identified by scATACseq. Treatment with the EZH2 specific inhibitor GSK343, blocked SMAD3-dependent cis co-accessibility and inhibited myofibroblast activation. This mechanism, through which TGFβ signals directly to chromatin, represents a critical determinant of fibrotic, differentiated states.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The differential role of Smad2 and Smad3 in the regulation of pro-fibrotic TGFbeta1 responses in human proximal-tubule epithelial cells.Biochem J. 2006 Jan 15;393(Pt 2):601-7. doi: 10.1042/BJ20051106. Biochem J. 2006. PMID: 16253118 Free PMC article.
-
Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs.Exp Mol Pathol. 2015 Jun;98(3):346-51. doi: 10.1016/j.yexmp.2015.03.033. Epub 2015 Mar 28. Exp Mol Pathol. 2015. PMID: 25828392 Free PMC article.
-
Epigenomic Evaluation of Cholangiocyte Transforming Growth Factor-β Signaling Identifies a Selective Role for Histone 3 Lysine 9 Acetylation in Biliary Fibrosis.Gastroenterology. 2021 Feb;160(3):889-905.e10. doi: 10.1053/j.gastro.2020.10.008. Epub 2020 Oct 12. Gastroenterology. 2021. PMID: 33058867 Free PMC article.
-
Identification of Predictive Markers for the Generation of Well-Differentiated Human Induced Pluripotent Stem Cell-Derived Kidney Organoids.Stem Cells Dev. 2021 Nov;30(22):1103-1114. doi: 10.1089/scd.2021.0197. Epub 2021 Nov 1. Stem Cells Dev. 2021. PMID: 34549597
-
Transforming growth factor-beta and Smad signalling in kidney diseases.Nephrology (Carlton). 2005 Feb;10(1):48-56. doi: 10.1111/j.1440-1797.2005.00334.x. Nephrology (Carlton). 2005. PMID: 15705182 Review.
Cited by
-
Nuclear ATP-citrate lyase regulates chromatin-dependent activation and maintenance of the myofibroblast gene program.Nat Cardiovasc Res. 2024 Jul;3(7):869-882. doi: 10.1038/s44161-024-00502-3. Epub 2024 Jul 5. Nat Cardiovasc Res. 2024. PMID: 39196175 Free PMC article.
-
YAP/TAZ: Molecular pathway and disease therapy.MedComm (2020). 2023 Aug 9;4(4):e340. doi: 10.1002/mco2.340. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37576865 Free PMC article. Review.
-
Fibrosis-the tale of H3K27 histone methyltransferases and demethylases.Front Cell Dev Biol. 2023 Jul 5;11:1193344. doi: 10.3389/fcell.2023.1193344. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37476157 Free PMC article. Review.
-
Advanced 3D imaging and organoid bioprinting for biomedical research and therapeutic applications.Adv Drug Deliv Rev. 2024 May;208:115237. doi: 10.1016/j.addr.2024.115237. Epub 2024 Mar 5. Adv Drug Deliv Rev. 2024. PMID: 38447931 Review.
References
-
- Ziyadeh FN, et al. Long-term prevention of renal insufficiency, excess matrix gene expression, and glomerular mesangial matrix expansion by treatment with monoclonal antitransforming growth factor-β antibody in db/db diabetic mice. Proc. Natl Acad. Sci. USA. 2000;97:8015–8020. doi: 10.1073/pnas.120055097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

