Organotypic cultures as aging associated disease models
- PMID: 36435511
- PMCID: PMC9740367
- DOI: 10.18632/aging.204361
Organotypic cultures as aging associated disease models
Abstract
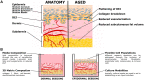
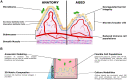
Aging remains a primary risk factor for a host of diseases, including leading causes of death. Aging and associated diseases are inherently multifactorial, with numerous contributing factors and phenotypes at the molecular, cellular, tissue, and organismal scales. Despite the complexity of aging phenomena, models currently used in aging research possess limitations. Frequently used in vivo models often have important physiological differences, age at different rates, or are genetically engineered to match late disease phenotypes rather than early causes. Conversely, routinely used in vitro models lack the complex tissue-scale and systemic cues that are disrupted in aging. To fill in gaps between in vivo and traditional in vitro models, researchers have increasingly been turning to organotypic models, which provide increased physiological relevance with the accessibility and control of in vitro context. While powerful tools, the development of these models is a field of its own, and many aging researchers may be unaware of recent progress in organotypic models, or hesitant to include these models in their own work. In this review, we describe recent progress in tissue engineering applied to organotypic models, highlighting examples explicitly linked to aging and associated disease, as well as examples of models that are relevant to aging. We specifically highlight progress made in skin, gut, and skeletal muscle, and describe how recently demonstrated models have been used for aging studies or similar phenotypes. Throughout, this review emphasizes the accessibility of these models and aims to provide a resource for researchers seeking to leverage these powerful tools.
Keywords: intestine; organotypic; skeletal muscle; skin; tissue engineering.
Conflict of interest statement
Figures

Similar articles
-
In vitro psoriasis models with focus on reconstructed skin models as promising tools in psoriasis research.Exp Biol Med (Maywood). 2017 Jun;242(11):1158-1169. doi: 10.1177/1535370217710637. Exp Biol Med (Maywood). 2017. PMID: 28585891 Free PMC article.
-
Decellularized tissues as platforms for in vitro modeling of healthy and diseased tissues.Acta Biomater. 2020 Jul 15;111:1-19. doi: 10.1016/j.actbio.2020.05.031. Epub 2020 May 25. Acta Biomater. 2020. PMID: 32464269 Review.
-
Using human epithelial amnion cells in human de-epidermized dermis for skin regeneration.J Dermatol Sci. 2016 Jan;81(1):26-34. doi: 10.1016/j.jdermsci.2015.10.018. Epub 2015 Oct 31. J Dermatol Sci. 2016. PMID: 26596214
-
Generation of Genetically Modified Organotypic Skin Cultures Using Devitalized Human Dermis.J Vis Exp. 2015 Dec 14;(106):e53280. doi: 10.3791/53280. J Vis Exp. 2015. PMID: 26709715 Free PMC article.
-
Advanced Organotypic In Vitro Model Systems for Host-Microbial Coculture.Biochip J. 2023 May 22:1-27. doi: 10.1007/s13206-023-00103-5. Online ahead of print. Biochip J. 2023. PMID: 37363268 Free PMC article. Review.
Cited by
-
Microphysiological systems for human aging research.Aging Cell. 2024 Mar;23(3):e14070. doi: 10.1111/acel.14070. Epub 2024 Jan 5. Aging Cell. 2024. PMID: 38180277 Free PMC article. Review.
-
3D Models Currently Proposed to Investigate Human Skin Aging and Explore Preventive and Reparative Approaches: A Descriptive Review.Biomolecules. 2024 Aug 26;14(9):1066. doi: 10.3390/biom14091066. Biomolecules. 2024. PMID: 39334833 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention. The State of Aging & Health in America 2013. Atlanta, GA: Centers for Disease Control and Prevention, US Dept of Health and Human Services. 2013. https://www.cdc.gov/aging/pdf/state-aging-health-in-america-2013.pdf.
-
- Gerteis J, Izrael D, Deitz D, LeRoy L, Ricciardi R, Miller T, Basu J. Multiple Chronic Conditions Chartbook. Agency for Healthcare Research and Quality. 2014.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

