Integration of single-cell transcriptomes and biological function reveals distinct behavioral patterns in bone marrow endothelium
- PMID: 36433940
- PMCID: PMC9700769
- DOI: 10.1038/s41467-022-34425-z
Integration of single-cell transcriptomes and biological function reveals distinct behavioral patterns in bone marrow endothelium
Abstract
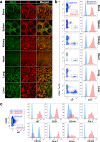
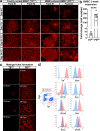
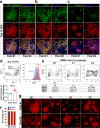
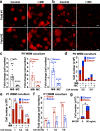
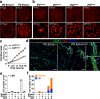
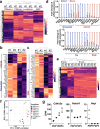
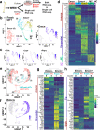
Heterogeneity of endothelial cell (EC) populations reflects their diverse functions in maintaining tissue's homeostasis. However, their phenotypic, molecular, and functional properties are not entirely mapped. We use the Tie2-CreERT2;Rosa26-tdTomato reporter mouse to trace, profile, and cultivate primary ECs from different organs. As paradigm platform, we use this strategy to study bone marrow endothelial cells (BMECs). Single-cell mRNA sequencing of primary BMECs reveals that their diversity and native molecular signatures is transitorily preserved in an ex vivo culture that conserves key cell-to-cell microenvironment interactions. Macrophages sustain BMEC cellular diversity and expansion and preserve sinusoidal-like BMECs ex vivo. Endomucin expression discriminates BMECs in populations exhibiting mutually exclusive properties and distinct sinusoidal/arterial and tip/stalk signatures. In contrast to arterial-like, sinusoidal-like BMECs are short-lived, form 2D-networks, contribute to in vivo angiogenesis, and support hematopoietic stem/progenitor cells in vitro. This platform can be extended to other organs' ECs to decode mechanistic information and explore therapeutics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A novel strategy for isolation of mice bone marrow endothelial cells (BMECs).Stem Cell Res Ther. 2021 May 3;12(1):267. doi: 10.1186/s13287-021-02352-3. Stem Cell Res Ther. 2021. PMID: 33941266 Free PMC article.
-
Spatial heterogeneity of bone marrow endothelial cells unveils a distinct subtype in the epiphysis.Nat Cell Biol. 2023 Oct;25(10):1415-1425. doi: 10.1038/s41556-023-01240-7. Epub 2023 Oct 5. Nat Cell Biol. 2023. PMID: 37798545 Free PMC article.
-
Generation of BMEC Lines and in vitro BMEC-HSPC Co-culture Assays.Bio Protoc. 2018 Nov 5;8(21):e3079. doi: 10.21769/BioProtoc.3079. eCollection 2018 Nov 5. Bio Protoc. 2018. PMID: 34532536 Free PMC article.
-
Soluble factors from bone marrow endothelial cells regulate differentiation and proliferation of hematopoietic and endothelial lineages and embryonic stem cells.Sheng Li Xue Bao. 2013 Aug 25;65(4):433-44. Sheng Li Xue Bao. 2013. PMID: 23963075 Review.
-
Regulation of hematopoiesis by microvascular endothelium.Leuk Lymphoma. 1997 Nov;27(5-6):375-86. doi: 10.3109/10428199709058305. Leuk Lymphoma. 1997. PMID: 9477120 Review.
Cited by
-
Bone marrow niches for hematopoietic stem cells.Hemasphere. 2024 Jul 31;8(8):e133. doi: 10.1002/hem3.133. eCollection 2024 Aug. Hemasphere. 2024. PMID: 39086665 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

