KARAJ: An Efficient Adaptive Multi-Processor Tool to Streamline Genomic and Transcriptomic Sequence Data Acquisition
- PMID: 36430895
- PMCID: PMC9694301
- DOI: 10.3390/ijms232214418
KARAJ: An Efficient Adaptive Multi-Processor Tool to Streamline Genomic and Transcriptomic Sequence Data Acquisition
Abstract
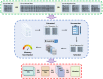
Here we developed KARAJ, a fast and flexible Linux command-line tool to automate the end-to-end process of querying and downloading a wide range of genomic and transcriptomic sequence data types. The input to KARAJ is a list of PMCIDs or publication URLs or various types of accession numbers to automate four tasks as follows; firstly, it provides a summary list of accessible datasets generated by or used in these scientific articles, enabling users to select appropriate datasets; secondly, KARAJ calculates the size of files that users want to download and confirms the availability of adequate space on the local disk; thirdly, it generates a metadata table containing sample information and the experimental design of the corresponding study; and lastly, it enables users to download supplementary data tables attached to publications. Further, KARAJ provides a parallel downloading framework powered by Aspera connect which reduces the downloading time significantly.
Keywords: Bioinformatics; Download; FASTQ; Genomics; Linux; biological data; sequence data; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
GeoBoost: accelerating research involving the geospatial metadata of virus GenBank records.Bioinformatics. 2018 May 1;34(9):1606-1608. doi: 10.1093/bioinformatics/btx799. Bioinformatics. 2018. PMID: 29240889 Free PMC article.
-
TCGA Expedition: A Data Acquisition and Management System for TCGA Data.PLoS One. 2016 Oct 27;11(10):e0165395. doi: 10.1371/journal.pone.0165395. eCollection 2016. PLoS One. 2016. PMID: 27788220 Free PMC article.
-
ILIAD: a suite of automated Snakemake workflows for processing genomic data for downstream applications.BMC Bioinformatics. 2023 Nov 8;24(1):424. doi: 10.1186/s12859-023-05548-x. BMC Bioinformatics. 2023. PMID: 37940870 Free PMC article.
-
grabseqs: simple downloading of reads and metadata from multiple next-generation sequencing data repositories.Bioinformatics. 2020 Jun 1;36(11):3607-3609. doi: 10.1093/bioinformatics/btaa167. Bioinformatics. 2020. PMID: 32154830 Free PMC article.
-
Pathogen metadata platform: software for accessing and analyzing pathogen strain information.BMC Bioinformatics. 2016 Sep 15;17(1):379. doi: 10.1186/s12859-016-1231-2. BMC Bioinformatics. 2016. PMID: 27634291 Free PMC article.
Cited by
-
A Comprehensive Investigation of Genomic Variants in Prostate Cancer Reveals 30 Putative Regulatory Variants.Int J Mol Sci. 2023 Jan 27;24(3):2472. doi: 10.3390/ijms24032472. Int J Mol Sci. 2023. PMID: 36768794 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

