Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer
- PMID: 36428736
- PMCID: PMC9688237
- DOI: 10.3390/cancers14225643
Mechanistic and Clinical Evidence Supports a Key Role for Cell Division Cycle Associated 5 (CDCA5) as an Independent Predictor of Outcome in Invasive Breast Cancer
Abstract
Background: Cell Division Cycle Associated 5 (CDCA5) plays a role in the phosphoinositide 3-kinase (PI3K)/AKT/mTOR signalling pathway involving cell division, cancer cell migration and apoptosis. This study aims to assess the prognostic and biological value of CDCA5 in breast cancer (BC).
Methods: The biological and prognostic value of CDCA5 were evaluated at mRNA (n = 5109) and protein levels (n = 614) utilizing multiple well-characterized early stage BC cohorts. The effects of CDCA5 knockdown (KD) on multiple oncogenic assays were assessed in vitro using a panel of BC cell lines.
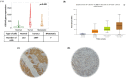
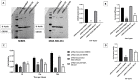
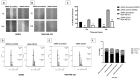
Results: this study examined cohorts showed that high CDCA5 expression was correlated with features characteristic of aggressive behavior and poor prognosis, including the presence of high grade, large tumor size, lymphovascular invasion (LVI), hormone receptor negativity and HER2 positivity. High CDCA5 expression, at both mRNA and protein levels, was associated with shorter BC-specific survival independent of other variables (p = 0.034, Hazard ratio (HR) = 1.6, 95% CI; 1.1-2.3). In line with the clinical data, in vitro models indicated that CDCA5 depletion results in a marked decrease in BC cell invasion and migration abilities and a significant accumulation of the BC cells in the G2/M-phase.
Conclusions: These results provide evidence that CDCA5 plays an important role in BC development and metastasis and could be used as a potential biomarker to predict disease progression in BC.
Keywords: CDCA5; breast cancer; prognosis; progression.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

Similar articles
-
CDCA5 functions as a tumor promoter in bladder cancer by dysregulating mitochondria-mediated apoptosis, cell cycle regulation and PI3k/AKT/mTOR pathway activation.J Cancer. 2020 Feb 10;11(9):2408-2420. doi: 10.7150/jca.35372. eCollection 2020. J Cancer. 2020. PMID: 32201512 Free PMC article.
-
CDCA5 promotes the progression of breast cancer and serves as a potential prognostic biomarker.Oncol Rep. 2022 Oct;48(4):172. doi: 10.3892/or.2022.8387. Epub 2022 Aug 25. Oncol Rep. 2022. PMID: 36004470 Free PMC article.
-
Upregulation of Cyclin B2 (CCNB2) in breast cancer contributes to the development of lymphovascular invasion.Am J Cancer Res. 2022 Feb 15;12(2):469-489. eCollection 2022. Am J Cancer Res. 2022. PMID: 35261781 Free PMC article.
-
The clinical significance of cyclin B1 (CCNB1) in invasive breast cancer with emphasis on its contribution to lymphovascular invasion development.Breast Cancer Res Treat. 2023 Apr;198(3):423-435. doi: 10.1007/s10549-022-06801-2. Epub 2022 Nov 22. Breast Cancer Res Treat. 2023. PMID: 36418517 Free PMC article.
-
PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell signaling to clinical trials.Pharmacol Ther. 2017 Jul;175:91-106. doi: 10.1016/j.pharmthera.2017.02.037. Epub 2017 Feb 16. Pharmacol Ther. 2017. PMID: 28216025 Review.
Cited by
-
The impact of CDCA5 expression on the immune microenvironment and its potential utility as a biomarker for PD-L1/PD-1 inhibitors in lung adenocarcinoma.Transl Oncol. 2024 Aug;46:102024. doi: 10.1016/j.tranon.2024.102024. Epub 2024 Jun 4. Transl Oncol. 2024. PMID: 38838437 Free PMC article.
-
CDCA5-EEF1A1 interaction promotes progression of clear cell renal cell carcinoma by regulating mTOR signaling.Cancer Cell Int. 2024 Apr 24;24(1):147. doi: 10.1186/s12935-024-03330-4. Cancer Cell Int. 2024. PMID: 38658931 Free PMC article.
-
CDCA5 accelerates progression of breast cancer by promoting the binding of E2F1 and FOXM1.J Transl Med. 2024 Jul 8;22(1):639. doi: 10.1186/s12967-024-05443-w. J Transl Med. 2024. PMID: 38978058 Free PMC article.
References
-
- Vijai J., Kirchhoff T., Schrader K.A., Brown J., Dutra-Clarke A.V., Manschreck C., Hansen N., Rau-Murthy R., Sarrel K., Przybylo J., et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet. 2013;9:e1003220. doi: 10.1371/journal.pgen.1003220. - DOI - PMC - PubMed
-
- Johanneson B., Deutsch K., McIntosh L., Friedrichsen-Karyadi D.M., Janer M., Kwon E.M., Iwasaki L., Hood L., Ostrander E.A., Stanford J.L. Suggestive genetic linkage to chromosome 11p11.2-q12.2 in hereditary prostate cancer families with primary kidney cancer. Prostate. 2007;67:732–742. doi: 10.1002/pros.20528. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

