MYB44-ENAP1/2 restricts HDT4 to regulate drought tolerance in Arabidopsis
- PMID: 36413574
- PMCID: PMC9681084
- DOI: 10.1371/journal.pgen.1010473
MYB44-ENAP1/2 restricts HDT4 to regulate drought tolerance in Arabidopsis
Abstract
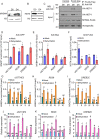
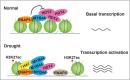
Histone acetylation has been shown to involve in stress responses. However, the detailed molecular mechanisms that how histone deacetylases and transcription factors function in drought stress response remain to be understood. In this research, we show that ENAP1 and ENAP2 are positive regulators of drought tolerance in plants, and the enap1enap2 double mutant is more sensitive to drought stress. Both ENAP1 and ENAP2 interact with MYB44, a transcription factor that interacts with histone deacetylase HDT4. Genetics data show that myb44 null mutation enhances the sensitivity of enap1enap2 to drought stress. Whereas, HDT4 negatively regulates plant drought response, the hdt4 mutant represses enap1enap2myb44 drought sensitive phenotype. In the normal condition, ENAP1/2 and MYB44 counteract the HDT4 function for the regulation of H3K27ac. Upon drought stress, the accumulation of MYB44 and reduction of HDT4 leads to the enrichment of H3K27ac and the activation of target gene expression. Overall, this research provides a novel molecular mechanism by which ENAP1, ENAP2 and MYB44 form a complex to restrict the function of HDT4 in the normal condition; under drought condition, accumulated MYB44 and reduced HDT4 lead to the elevation of H3K27ac and the expression of drought responsive genes, as a result, plants are drought tolerant.
Copyright: © 2022 Zhao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress.Int J Mol Sci. 2014 Feb 13;15(2):2517-37. doi: 10.3390/ijms15022517. Int J Mol Sci. 2014. PMID: 24531138 Free PMC article.
-
The FBA motif-containing protein AFBA1 acts as a novel positive regulator of ABA response in Arabidopsis.Plant Cell Physiol. 2017 Mar 1;58(3):574-586. doi: 10.1093/pcp/pcx003. Plant Cell Physiol. 2017. PMID: 28184867
-
Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis.J Exp Bot. 2016 Mar;67(6):1703-13. doi: 10.1093/jxb/erv562. Epub 2016 Jan 4. J Exp Bot. 2016. PMID: 26733691
-
Tight interconnection and multi-level control of Arabidopsis MYB44 in MAPK cascade signalling.PLoS One. 2013;8(2):e57547. doi: 10.1371/journal.pone.0057547. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437396 Free PMC article.
-
A histone deacetylase, GhHDT4D, is positively involved in cotton response to drought stress.Plant Mol Biol. 2020 Sep;104(1-2):67-79. doi: 10.1007/s11103-020-01024-9. Epub 2020 Jul 3. Plant Mol Biol. 2020. PMID: 32621165
Cited by
-
Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression.Genes (Basel). 2023 May 30;14(6):1199. doi: 10.3390/genes14061199. Genes (Basel). 2023. PMID: 37372378 Free PMC article.
-
The chromosome-level genome of the submerged plant Cryptocoryne crispatula provides insights into the terrestrial-freshwater transition in Araceae.DNA Res. 2024 Feb 1;31(1):dsae003. doi: 10.1093/dnares/dsae003. DNA Res. 2024. PMID: 38245835 Free PMC article.
-
Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response.Sci Adv. 2024 Jul 26;10(30):eado2825. doi: 10.1126/sciadv.ado2825. Epub 2024 Jul 26. Sci Adv. 2024. PMID: 39058774 Free PMC article.
-
Genomic survey of MYB gene family in six pearl millet (Pennisetum glaucum) varieties and their response to abiotic stresses.Genetica. 2023 Jun;151(3):251-265. doi: 10.1007/s10709-023-00188-8. Epub 2023 Jun 2. Genetica. 2023. PMID: 37266766
-
Plant responses to abiotic stress regulated by histone acetylation.Front Plant Sci. 2024 Jul 16;15:1404977. doi: 10.3389/fpls.2024.1404977. eCollection 2024. Front Plant Sci. 2024. PMID: 39081527 Free PMC article. Review.
References
-
- Yang X, Lu M, Wang Y, Wang Y, Liu Z, Chen S. Response mechanism of plants to drought stress. Horticulturae. 2021;7(3):50.
-
- Kaur G, Asthir B. Molecular responses to drought stress in plants. Biologia Plantarum. 2017;61(2):201–9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

