Complex host/symbiont integration of a multi-partner symbiotic system in the eusocial aphid Ceratovacuna japonica
- PMID: 36404929
- PMCID: PMC9672956
- DOI: 10.1016/j.isci.2022.105478
Complex host/symbiont integration of a multi-partner symbiotic system in the eusocial aphid Ceratovacuna japonica
Abstract
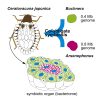
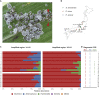
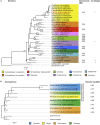
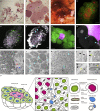
Some hemipteran insects rely on multiple endosymbionts for essential nutrients. However, the evolution of multi-partner symbiotic systems is not well-established. Here, we report a co-obligate symbiosis in the eusocial aphid, Ceratovacuna japonica. 16S rRNA amplicon sequencing unveiled co-infection with a novel Arsenophonus sp. symbiont and Buchnera aphidicola, a common obligate endosymbiont in aphids. Both symbionts were housed within distinct bacteriocytes and were maternally transmitted. The Buchnera and Arsenophonus symbionts had streamlined genomes of 432,286 bp and 853,149 bp, respectively, and exhibited metabolic complementarity in riboflavin and peptidoglycan synthesis pathways. These anatomical and genomic properties were similar to those of independently evolved multi-partner symbiotic systems, such as Buchnera-Serratia in Lachninae and Periphyllus aphids, representing remarkable parallelism. Furthermore, symbiont populations and bacteriome morphology differed between reproductive and soldier castes. Our study provides the first example of co-obligate symbiosis in Hormaphidinae and gives insight into the evolutionary genetics of this complex system.
Keywords: Evolutionary biology; Genetics; Molecular biology.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Compartmentalized into Bacteriocytes but Highly Invasive: the Puzzling Case of the Co-Obligate Symbiont Serratia symbiotica in the Aphid Periphyllus lyropictus.Microbiol Spectr. 2022 Jun 29;10(3):e0045722. doi: 10.1128/spectrum.00457-22. Epub 2022 Jun 1. Microbiol Spectr. 2022. PMID: 35647657 Free PMC article.
-
Reinventing the Wheel and Making It Round Again: Evolutionary Convergence in Buchnera-Serratia Symbiotic Consortia between the Distantly Related Lachninae Aphids Tuberolachnus salignus and Cinara cedri.Genome Biol Evol. 2016 May 22;8(5):1440-58. doi: 10.1093/gbe/evw085. Genome Biol Evol. 2016. PMID: 27190007 Free PMC article.
-
Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts.Mol Ecol. 2017 Apr;26(8):2363-2378. doi: 10.1111/mec.13910. Epub 2016 Dec 19. Mol Ecol. 2017. PMID: 27862540
-
Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum.C R Biol. 2009 Nov;332(11):1034-49. doi: 10.1016/j.crvi.2009.09.007. Epub 2009 Oct 14. C R Biol. 2009. PMID: 19909925 Review.
-
Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont.Cell Mol Life Sci. 2011 Apr;68(8):1297-309. doi: 10.1007/s00018-011-0645-2. Epub 2011 Mar 10. Cell Mol Life Sci. 2011. PMID: 21390549 Free PMC article. Review.
Cited by
-
The nutritional dimension of facultative bacterial symbiosis in aphids: Current status and methodological considerations for future research.Curr Res Insect Sci. 2023 Dec 20;5:100070. doi: 10.1016/j.cris.2023.100070. eCollection 2024. Curr Res Insect Sci. 2023. PMID: 38222793 Free PMC article. Review.
-
16S rRNA Gene Sequencing of Six Psyllid Species of the Family Carsidaridae Identified Various Bacteria Including Symbiopectobacterium.Microbes Environ. 2023;38(3):ME23045. doi: 10.1264/jsme2.ME23045. Microbes Environ. 2023. PMID: 37612118 Free PMC article.
-
microRNA maintains nutrient homeostasis in the symbiont-host interaction.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2406925121. doi: 10.1073/pnas.2406925121. Epub 2024 Aug 28. Proc Natl Acad Sci U S A. 2024. PMID: 39196627
-
Division of labor within psyllids: metagenomics reveals an ancient dual endosymbiosis with metabolic complementarity in the genus Cacopsylla.mSystems. 2023 Oct 26;8(5):e0057823. doi: 10.1128/msystems.00578-23. Epub 2023 Sep 28. mSystems. 2023. PMID: 37768069 Free PMC article.
-
PacBio Hi-Fi genome assembly of Sipha maydis, a model for the study of multipartite mutualism in insects.Sci Data. 2024 May 4;11(1):450. doi: 10.1038/s41597-024-03297-x. Sci Data. 2024. PMID: 38704391 Free PMC article.
References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Aoki S., Kurosu U. Psyche (Stuttg); 2010. A Review of the Biology of Cerataphidini (Hemiptera, Aphididae, Hormaphidinae), Focusing Mainly on Their Life Cycles, Gall Formation, and Soldiers.
-
- Ayoubi A., Talebi A.A., Fathipour Y., Mehrabadi M. Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae) Insect Sci. 2020;27:86–98. - PubMed
-
- Baumann P., Baumann L., Lai C.Y., Rouhbakhsh D., Moran N.A., Clark M.A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 1995;49:55–94. - PubMed
LinkOut - more resources
Full Text Sources

